A new function for sleep
Sleep has a primary role on human life. We spend 1/3 of our life sleeping, but the real complete function of sleep hasn’t been discovered yet. The research team of D. Zada et al. at Bar-Ilan University, Israel [1] , tried to find a correlation between sleep, chromosome dynamics and double strand breaks. In particular, their hypothesis was that night sleep time in neurons is fundamental to repair the DSBs generated by neuronal and metabolic activity during wakefulness. Exploiting the transparence of zebrafish larvae, it has been possible for them to study the chromosome dynamics by imaging with fluorescent proteins, confirming that it increases significantly during the night. In fact, in zebrafish larvae sleep follows circadian rhythms (14 hours/10 hours). Sleep resulted to be a sufficient factor to increase chromosome dynamics: after treating larvae with melatonin for 3 hours, scientists observed that sleep time increased and successively also chromosome dynamics. At this point, they created a mutant form of zebrafish larvae deprived of aanat2, the gene for the last enzyme of melatonin biosynthesis. This mutant lacked the signaling of melatonin, so sleep time decreased and consequentially also chromosome dynamics, confirming that sleep is also necessary to increase chromosome dynamics.
The next step for the scientists was to quantify the amount of double strand breaks in neurons. One possible marker of DSBs is γH2AX, that was quantified by immunostaining. The results obtained brought the team to confirm the correlation between sleep, chromosome dynamics, and double strand breaks: induction of night sleep time increases chromosome dynamics that acts on DSBs, repairing them. Despite sleep is composed of different phases, all with different characteristics, neuronal activity is on average low. The last experiments have been focused on correlation between neuronal activity and chromosome dynamics. Through optogenetic studies, the authors pointed out that in mutant larvae for Chr2 (Chr2+) stimulated with blue light (to increase neuronal activity) the level of chromosome dynamics decreased. This correlation is negative but moderate and it has been confirmed also through the treating of larvae with BAPTA-AM, inhibiting neuronal activity and finding out that it isn’t directed correlated to chromosome dynamics, but it can affect it via DBSs. The last experiment has been conducted in reverse respect to the others discussed until now: the scientists induced DSBs using ETO, and they discovered that their repair occurs only after the increase of chromosome dynamics induced by sleep. To sum up, the experiments conducted by the team demonstrated that probably is the reaching of a threshold number of DSBs that induces sleep that in turn is the factor responsible of the increasing of chromosome dynamics. But why does chromosome dynamics increase during sleep? Without it the regulation of different key process in the nucleus, like epigenetic, transcription and interaction between promoter and enhancers, cannot occur. Future studies need to be focused on the characterization of markers and analysis of what happens in the nucleus: which genes are transcribed? What kind of epigenetic modifications occur? Which interactions of what promoters and enhancers are stimulated?
However, the article is well structured, and all the experiments are complete and explained in a clear way, with tests and counterchecks that validate the hypothesis. Furthermore, some experiments have been taken also on Schwann’s and endothelial cells, obtaining results that suggest that the kind of correlation searched from the team is true only in excitable cells.
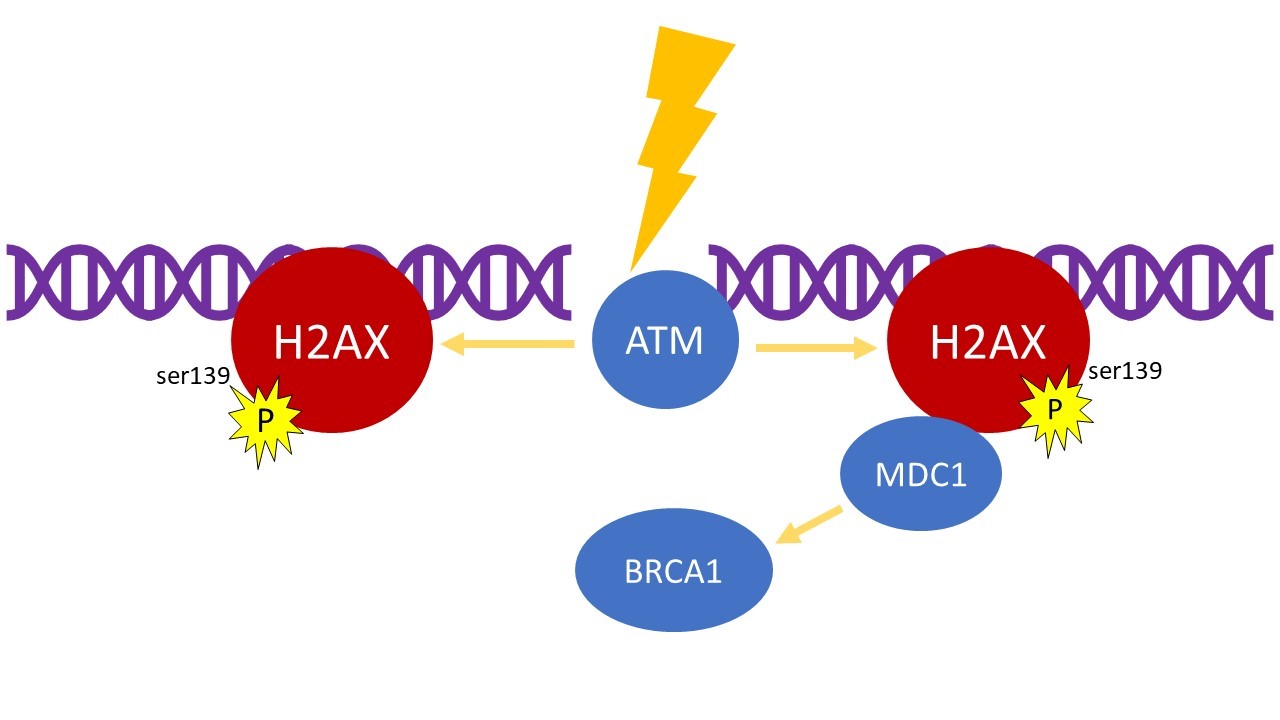
Fig. 1 H2Ax as a proxy for double strand break. H2AX is phosphorylated (γH2AX) on serine 139 by a kinase, and this process mediate the arrival of the protein MCD1 on the break site of the double helix. This latter recruit other proteins that amplify the signal permitting the efficient repair of DSBs by other factors like BRCA1.
One of the weak points of the study is that the experiments have been conducted on single neurons but not in neural networks, that could respond in a different way. Moreover, larvae used have been defined “sleepy” only by behavioral aspects. In fact, sleep in zebrafish is defined by at least one minute of immobility. Toward this, the characterization of molecular “sleep markers” could be an important factor for the validation of this kind of experiments. Moreover, knowing the different phases of sleep, it would be appropriate to relate chromosome dynamics to different phases of EEG.
Besides, there’s another controversial issue…
The phosphorylation of histone H2AX has been used as a marker for DSBs during several years. There is a multitude of papers that confirm the efficiency of the method. However, some new studies called into question this method. Some researchers assume that the quantification through γH2AX overestimates the real number of DSBs, because the phosphorylation of H2AX seems to be a factor that characterize not only DSBs but also SSBs or other processes like chromatin remodeling or apoptosis induction. On the other hand, some others think that this marker underestimates the amount of DSBs, pointing out that using the antibodies to visualize γH2AX find them as foci only at sites where the active DSB repair process is active. To further validate the hypothesis, quantify the number of DSBs using more than one method could have given the paper an extra point. For example, DSBs can be quantified using a TdT (Terminal deoxynucleotidyl Transferase). TdT mark DSBs introducing a dATP radiolabeled on 3’-OH. This can be done after a process of NGR (Nick and Gap Repair), to be sure to label only DSBs.
References
- Zada, I. Bronshtein, T. Lerer-Goldshtein, Y. Garini & L. Appelbaum. Sleep increases chromosome dynamics to enable reduction of accumulating DNA damage in single neurons. Nature Communications volume10, Article number: 895 (2019)
- Emmy P. Rogakou, Duane R. Pilch, Ann H. Orr, Vessela S. Ivanova, and William M. Bonner. DNA Double-stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139*. THE JOURNAL OF BIOLOGICAL CHEMISTRY 273, No. 10, Issue of March 6, pp. 5858–5868, 1998
- Yingjie Zhu, Anna Biernacka, Benjamin Pardo, Norbert Dojer, Romain Forey, Magdalena Skrzypczak, Bernard Fongang, Jules Nde, Razie Yousefi, Philippe Pasero, Krzysztof Ginalski & Maga Rowicka. qDSB-Seq is a general method for genome-wide quantification of DNA double-strand breaks using sequencing. Nature Communications volume10, Article number: 2313 (2019)
- Somaira Nowsheen, Khaled Aziz, Kuntian Luo, Min Deng, Bo Qin, Jian Yuan, Karthik B. Jeganathan, Jia Yu, Henan Zhang, Wei Ding, Jan M. van Deursen & Zhenkun Lou. ZNF506-dependent positive feedback loop regulates H2AX signaling after DNA damage. Nature Communications volume9, Article number: 2736 (2018)
- Marie-Chantal Grégoire, Julien Massonneau, Frédéric Leduc, Mélina Arguin,Marc-André Brazeau, Guylain Boissonneault. Quantification and genome-wide mapping of DNA double-strandbreaks. DNA Repair 48 (2016) 63–68
