Bad news for HIV
- ARTs are not able to eradicate the HIV from reservoir
- ARTs may have toxic effects on the organism
- Treatment with ARTs continue lifelong
Therefore, ARTs are able to stop just HIV replication. Instead, the aim of the new studies is to destroy hidden reservoirs of HIV, that is fundamental to cure the disease. There are new drugs, called LRAs, tested to wake up the reservoirs. These drugs are used in combination with ARTs in a strategy called “shock and kill” [2], where LRAs (for example Vorinostat) induce the disrupting of latency and ARTs lead to death of HIV-producing cells [2]. LRAs strike some signaling pathways involved in maintenance of latency. The main pathway is the protein kinase C (PKC)-NF-κB pathway.
In the article “HIV latency is reserved by ACSS2-driven histone crotonylation” Jiang et al. [3] have found a particular epigenetic modification on the histone tails which reverses virus latency, without intervening on the signaling pathways illustrated above. This modification is called crotonylation (Figure 1) of the HIV long terminal repeat, a critical site for the establishment of the latent reservoir. A particular enzyme, acyl-CoA synthetase short-chain family member 2 (ACSS2) mainly present in gut tract, can add the crotonyl group to side chain of a lysine, component of the histone tails, inducing reactivation of HIV transcription.
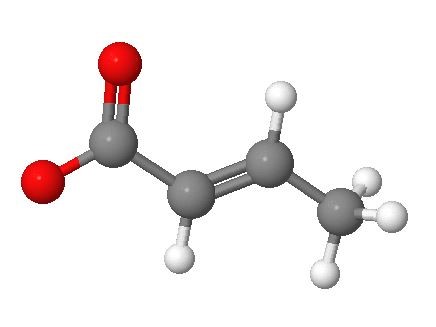
Figure 1. Molecular structure of the crotonate (performed with www.molcalc.org). The oxygen atom linked with a single bond has a negative charge.
In the first part of the study the authors sought to demonstrate that ACSS2 is responsible of the crotonylation of histone tails and this modification effectively leads to reactivation of HIV transcription. These experiments were made in vitro and ex vivo (in primary and resting CD4+ T cells). The results are in agreement with the hypothesis. It is now necessary verify the molecular mechanism of ACSS2-induced disrupting HIV latency. From the data it is clear that ACSS2 involves a global reprogramming of local chromatin at HIV LTR. Despite that LRAs and ACSS2 use two different targets to reactivate latent HIV, the authors proved to determine whether they have a synergistically effect. PEP005 (a LRA activating the PKC/NF-kB pathway) showed the most efficient synergy with ACSS2, also in primary CD4+ T cells. In order to have ACSS2-induced crotonylation really effective, it is important that the reactivation of latent HIV do not impact the level of immune activation. It was demonstrated that ACSS2 does not cause activation or suppression of T cells. Finally, the authors found that ACSS2 expression in vivo was increased by HIV infection during acute stage whereas it was decreased during chronic stage. These results derivate by studies on SIV-infected rhesus macaques. Furthermore, since ACSS2 is involved in lipid and fatty acid metabolism, it was sought if the modulation of this metabolism may regulate HIV replication. Effectively, during SIV infection lipid and fatty acid metabolism result altered but involvement in gut mucosal damage needs to be investigated.
In conclusion there are bad news for HIV but good news for patients infected by the virus because this research opens new possibilities for HIV cure. It is clear that common anti-retroviral therapies are essentials for the removal of the virus and they will still be used. Findings about intervention of chromatin reprogramming on maintenance of HIV latency and in particular the possibility to use jointly inducing crotonylation factors and LRAs allow to eradicate also HIV reservoirs. Future perspectives for therapeutic applications include the use of a method that can increase the activity of ACSS2 in order to increase histone crotonylation and to break the latency of the virus.
References
- Flint, V. Racaniello, G. Rall, A.M. Skalka, Principles of virology, volume II: Pathogenesis and Control, 4th edition, Washington, American Society of Microbiology, 2015.
- G. Deeks, HIV: Shock and kill, News and Views, Nature, vol. 487, 26 July 2012, p. 439-440
- Jiang, D. Nguyen et al. HIV latency is reversed by ACSS2-driven histone crotonylation. The Journal of clinical investigation.2018;128(3):1190:1198