Chromatin interactions in-situ analysis by CAPTURE
The article by Liu X. et al titled “In Situ Capture of Chromatin Interactions by Biotinylated dCas9” [1] presents a novel technique which allows to identify the actors in specific chromatin interactions such as locus-specific chromatin-regulating protein complexes and long-range DNA interactions. CAPTURE (CRISPR affinity purification in situ of regulatory elements) method is a CRISPR-Cas9 [2]-based affinity technique which employs a deactivated Cas9 nuclease driven on a specific DNA sequence by a short-guide RNA (sgRNA) [3][4]. The deactivation of cleavage activity on Cas9 is obtained via a point mutation of two amino-acids located in recognition and cleaving domain [5]. This dCas9 contains also a biotin-acceptor site which is used to purify and enrich the protein after its binding with the region of interest. As said before, the sgRNA has the role of guiding the Cas9 on-target; after the sequence-specific recognition between sgRNA-Cas9 complex and the target sequence, a biotin-ligase (Bir-A) operate a biotinylation on Cas9 acceptor site.
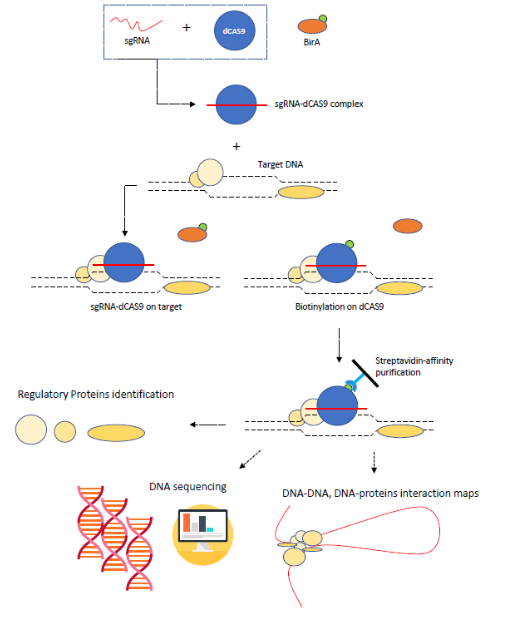
The formed complex – biotinylated dCas9, sgRNA and DNA – is then crosslinked with formaldehyde which is used to convert weak bonds in covalent bonds; the crosslinking interests also all the other molecules interacting in that site. As a result, an agglomerate of regulatory proteins and DNA sequences is obtained after cell lysis, sonication and DNA extraction. Another key-component of CAPTURE is streptavidin-biotin affinity purification system, which is made to target specific biotinylated targets like, in this case, the dCas9 in the complex of interest. The sample, which contains proteins complexes, is then reverse-crosslinked and ready for further analysis like HRMS (High Resolution Mass Spectrometry) proteomics and targeted-DNA sequencing. Analytical step is then moved in two directions: a) proteomic identification of regulators interacting with CREs (sequences of DNA which act as binding-domains for regulatory proteins), b) mapping of chromatin-chromatin long-distance interaction via paired end-sequencing. Proteomic and comparing analysis of different CREs is made by HPLC-MS (liquid chromatography – mass spectrometry) tools which allow the operator to detect any minimal mass shift in the sample with high accuracy and sensitivity. Comparison between more samples is made possible by iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) technique which uses different tags differing by a shift of mass. Coupled with liquid chromatography separation, iTRAQ system is capable to submit to MS (mass spectrometry) analysis the same proteins coming from different samples giving the scientist an overview of their relative abundance.
Otherwise, by processing the sample in a slightly different way, CAPTURE is capable of detecting long-range chromatin interaction giving informations about the chromatin folding. After the cross-linking, DNA is digested with restriction enzymes and submitted to paired end sequencing.
“Paired-end DNA Sequencing”®️ is used to restore the blunted DNA ends coming from digestion of the sample and sequence the regions. Data are then used to create maps of the protein – sequence or sequence – sequence interactions. The system provides an unbiased and specific analysis of interactions by RNA-specific guidance despite previously used antibody-based methods such Chromatin immunoprecipitation (ChIP), switching the target from a protein epitope to a DNA sequence. CAPTURE has the goal of overcoming previously used ChIP methods which suffer of a lower specificity and a low on-target ratio. In addition, ChIP based-methods need an “a priori” knowledge of the target protein. Isolation and enrichment by ChIP methods uses immunoreceptors which are mostly affected by aggressive pH, temperature and solvent variations. Quite the opposite CAPTURE count on a very robust streptavidin-biotin affinity purification method which is really strong and specific, resisting also to non-mild conditions and making the bond reverting sometimes a bit difficult. As a result, CAPTURE method, compared to FLAG ChIP-Seq and Cas9 immunoaffinity, shows a better enrichment and a lower off-targets ratio due to: a more specific-sequence recognition and a better affinity between streptavidin and biotin.
The nature of specific binding between biotin and streptavidin shows the need of a previous identification of all the endogenous proteins which are or biotinylated or have an affinity for streptavidin. These “high confidence proteins”, must be kept as a normalisation term and used as standard in measuring data. CAPTURE’s capabilities in locus specificity and sequence recognition were confirmed via targeting, by specific sgRNA, some telomeric sequences and β-globin gene enhancers (HS1-4) and promoters (HBG1-2 and HBB). The results proved a good and specific targeting of genomic sequences (coupling CAPTURE with sequencing of captured strands). The only exception found in genomic targeting was the unspecific enrichment of two high-homology sequences (HBG1 and HBG2) which weren’t discerned by the guided dCas9. The proteomic approach led to the enrichment of many proteins known to be acting on those sites like TERF-2 acting on telomeres and various regulation and transcription proteins acting on β-globin cluster’s CREs. Also new non-expected CREs associated proteins were found and validated via a functional RNA interference approach. The last application of CAPTURE, namely CAPTURE 3C (Chromosome Conformation Capture), showed astonishing results in the mapping of chromatin-chromatin interactions. Compared to 3C, 4C, 5C and ChIA-PET (Chromatin Interaction Analysis by Paired-End Tag Sequencing®️) the method is much more specific to a single sequence for its guidance nature since it is RNA-guided and not immunoreceptor-guided.
An experiment led on the β-globin cluster merging CAPTURE proteomics and CAPTURE 3C showed the presence of a major interaction and regulation hub in the HPFH (hereditary persistence of fetal hemoglobin) disease-associated locus. In conclusion CAPTURE technique offers a high resolution and specificity in locus-specific proteome and interactome analysis, non-depending on a “a priori” knowledge of the target locus or the proteins interacting with. CAPTURE is surely a very robust and valid alternative to current methods, with 2 huge advantages: 1) it acts on the sequence to find interactions (working backwards in comparison to older immunoaffinity-based techniques) 2) has a strong versatility which resides in the nature of the guiding sgRNA since it can be easily designed via online tools like “Optimized CRISPR Design” [2]. However for a good in-situ insight, the sgRNA must be chosen for a perfect match with the target sequence, to overcome this issue multiplexed sgRNAs for each region can be used [2]. The limitation of the system is represented by the necessary presence of a specific sequence, called PAM, located in the immediate vicinity of the target sequence. [6] For partial denaturing, the PAM sequence allows the attachment and recognition of the target DNA sequence by sgRNA-dCas9 complex.
References
- Liu, X.; Zhang, Y.; Chen, Y.; Li, M.; Zhou, F.; Li, K.; Cao, H.; Ni, M.; Liu, Y.; Gu, Z.; Dickerson, K. E.; Xie, S.; Hon, G. C.; Xuan, Z.; Zhang, M. Q.; Shao, Z.; and Xu, J. In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell, (170), 1028–1043. (2017).
- Ran, F. A.; Hsu, P. D.; Wright, J.; Agarwala, V.; Scott, D. A.; & Zhang F.; Genome engineering using the CRISPR-Cas9 system. Nature Protocols, 11(8), 2281-2308. (2013).
- Deltcheva, E.; Chylinski, K.; Sharma, C. M.; & Gonzales, K. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature, (471), 602-609. (2011).
- Hsu, P. D.; Scott, D. A.; Weinstein, J. A.; Ran, F. A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E. J.; Wu, X.; Shalem, O.; Cradick, T. J.; Marraffini, L. A.; Bao, G.; & Zhang, F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology, 9 (31), 827- 834. (2013).
- Nishimasu, H.; Ran, F.; Hsu, P. D.; Konermann, S.; Shehata, S. I.; Dohmae, N.; . . . Nureki, O.Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell, (156), 935-949. (2014).
- Deveau, H.; Barrangou, R.; Garneau, J. E.; Labonté, J.; & Fremaux, C. Phage Response to CRISPR-Encoded Resistance in Streptococcus thermophilus. Journal of Bacteriology, 190(4), 1390-1400. (2008).

