Reprogramming to Pluripotency Through CRISPR-based Chromatin Remodeling
In the last few years stem cells turned out to be a powerful tool for biological and medical field, in particular in regenerative medicine. Stem cells are undifferentiated cells able to, under certain stimuli, differentiate into many types of cells of the entire organism, both during early stage and growth process. Given their differentiating power, they act as an internal repair system in various tissues through replacing damaged or dead cells. The research has been focused in particular on embryonic stem cells (ESCs), classified as pluripotent cells due to their ability to differentiate into any cell of the three embryonic germ layers. However, using stem cells for research purposes has always been related to ethical problems: they are located in the blastocyst, so it is necessary to damage and destroy the early embryo in order to obtain ESCs.
In 2006, Yamanaka and collaborators succeeded in reprogramming differentiated somatic cells into induced pluripotent stem cells (iPSCs) through the ectopic expression of the transcriptional factors Oct4, Sox2, Klf4 and c-Myc (OSKM) [1]. These factors have been identified as Yamanaka’s reprogramming factors, playing a pivotal role to establish pluripotent regulatory network. ESCs and iPSCs are similar from many points of views: pluripotency network genes expression, protein levels, time of duplication, shape, and appearance. Nevertheless, iPSCs show some features of somatic cells from which they derive and chromatin seem to be more compact compared to ESCs. Although these findings led to important progress, it is largely unknown what kind of precise remodeling events on endogenous chromatin trigger reprogramming towards pluripotency. First of all, it is not clear whether the remodeling of a large number of pluripotency-related loci at the same time is necessary for iPSC induction.
CRISPR-Cas9 technology has rapidly changed the landscape to study and manipulate the genome. Derived from the bacterial immune system for cleaving foreign DNA, this technology consists of Cas9 endonuclease and a target-identifying CRISPR RNA. This latter is composed by two RNA engineered into a chimeric single-guide RNA (sgRNA), simplifying its use, that base pairs with the DNA target and can be easily programmed to recognized specific sequences [3].
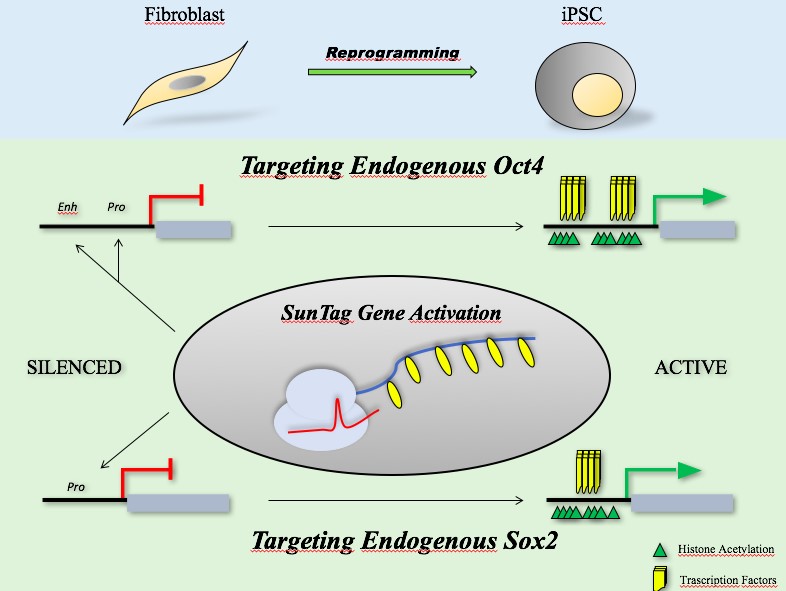
In this study, Liu and collaborators used a modified CRISPR technique, named CRISPR activation (CRISPRa), based on a deactivated Cas9 (dCas9). In order to convert Cas9 from a DNA scissor into a gene activator, it is necessary to disrupt its nuclease activity, converting the enzyme into a generic RNA-guided DNA-binding protein [3]. It has been fused with transcriptional activator domain able to recruit transcriptional machinery and acting as a pioneer factor to target silenced chromatin locus and promote transcription. To enhance the transcription, more transcriptional activator domains were linked to a protein scaffold fused with dCas9, system called SunTag technology [4].
Previous studies revealed that the activation of Sox2 could be a crucial event for the pluripotency induction, but there is no direct evidence of whether pluripotency can be induced by precise remodeling of Sox2 locus. At first, the authors showed that CRISPRa acted as a transcriptional activation of specific silenced gene loci. Furthermore, they found that the sole activation of Sox2 promoter induced the expression of other key genes involved in the pluripotency network and was sufficient to generate iPSC lines, ahough with low efficiency and high variability among the experiments [2]. The further activation of Oct4 promoter and enhancer, together with Sox2 promoter, increased the efficiency in the reprogramming process. So, there is a cooperativity of multiple gene loci in pluripotency induction.
In addition, to verify transcriptional activation, the authors analysed the epigenetic landscape remodeling at specific gene loci. Still using CRISPRa technology, the transcriptional activator domain has been replaced with an acetyl-transferasic domain, able to increase the acetylation level of targeted gene loci. The enhancement at Oct4 promoter and enhancer surprisingly led to the generation of iPSC lines, showing how changes of the epigenetic landscape is important and required for the pluripotency induction, proving new possible way to generate iPSC [2]. Compared to previous works, it was here demonstrated that pluripotency induction is possible by using chromatin remodeling or gene activation through CRISPRa technology, while Yamanaka et al. needed the ectopic expression of genes coding for key transcriptional factors of the pluripotency network.
Transducing exogenous genes through viral vectors evince potential problems related to transduction process itself and risk factors of external DNA introduction. Therefore, modulating epigenetic landscape in order to change cell fate is a crucial step for future researches, because it does not represent a drastic or excessively intrusive event towards the cell. Novel studies for iPSC reprograming employ mRNA and episomal plasmids carried by non-integrating viruses, easily to remove using specific antibodies. Problems related with these technics are the high costs that limit their applications.
Key genes in pluripotency induction are pivotal in maintaining the equilibrium during cell life. As a matter of fact different levels of transcriptional factors are characteristic of many steps during cell cycle. Uncontrolled variation of these latter could lead to tumor development, since some of the genes coding for Yamanaka reprogramming factors are identified as oncogenes. Another main turning point for future researches is to try to establish the pluripotency network using specific molecules able to reactivate and enhance silenced genes rather than inducing genomic changes or through ectopic genes expression. Although big steps have already been taken, in a future perspective it would be useful to enhance the reprogramming efficiency, which is at very low levels, in order to obtain process optimization.
References
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors, Cell, 2006 Aug 25;126(4):663-76, doi: 10.1016/j.stem.2017.12.001
- Peng Liu, Meng Chen, Yanxia Liu, Lei S. Qi, Sheng Ding. CRISPR-Based Chromatin Remodeling of the Endogenous Oct4 or Sox2 Locus Enables Reprogramming to Pluripotency, Cell Stem Cell, 2018 Feb 1;22(2):252-261, doi: 10.1016/j.stem.2017.12.001
- La Russa MF, Qi LS. The New State of the Art: Cas9 for Gene Activation and Repression, Mol Cell Biol, 2015 Nov;35(22):3800-9. doi: 10.1128/MCB.00512-15
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging, Cell, 2014 Oct 23;159(3):635-46. doi: 10.1016/j.cell.2014.09.039