CRISPR/Cas9 – mediated genome editing: a promising strategy to treat haemophilia B in newborn mice
Haemophilia B is a X-linked genetic disorder caused by mutations in coagulation factor IX gene, F9 gene. Because of the ineffective coagulative cascade, patients are subjected to easy bruising, severe bleeding and joint, muscle and intracranial haemorrhages. Today the most widely applied treatment for this disease is a replacement therapy with a recombinant form of factor IX [1], which still shows some limitations. This recombinant factor makes up for the lack of the natural one, but its therapeutic effect can be compromised by inhibitory alloantibodies developed by the organism of the patient. Also, it must be said that this treatment does not provide a cure for the pathology, neither does it prevent the onset of the symptoms. Furthermore, the technology required to produce recombinant factors is remarkably expensive and the treatment can rarely be adopted as prophylactic. Finding a definitive cure for this pathology has become a central point since 2000. A direct intervention on the gene of interest seems to be feasible as haemophilia is a monogenic disease, F9 gene is structurally simple and not tightly regulated and several studies demonstrated that an increase of 1-2% in clotting factor levels is sufficient to achieve an improvement in clinical conditions. An accurate correction of the mutated gene would guarantee a continuous endogenous expression of the lacking factor, solving the problem at its roots. Well characterized animal models (murine and canine) are already available and encourage further research in this field.
Vector-based gene therapy has been the first explored technique. It employs a viral or non-viral vector able to transfer the intact form of F9 into the target liver cells of the patient. Adenoassociated viruses (AAVs) were shown to be the best viral vectors thanks to their efficiency in transferring the gene and to their low immunogenicity [2, 3]. To restore gene activity, an alternative that has been rising interest in the last years involves the genome editing techniques, with accent on CRISPR/Cas9. CRISPR/Cas systems are bacterial adaptive immune systems based on a nuclease guided by a small RNA on a specific target DNA. The nuclease Cas9 induces a double strand break (DSB) on the target DNA and the DSB is then repaired by homologous direct repair (HDR) or non-homologous end joining repair (NHEJ). CRISPR/Cas system is guided to its genomic targets by an RNA-guided sequence, called sgRNA. By changing the sgRNA sequence, CRISPR/Cas9 system can be programmed to target virtually any DNA sequence of interest in the genome. Moreover, by introducing a specific DNA template for the HDR it is possible to obtain the desired corrected form of the gene [4, 5].
As the efficiency of Cas9 is considerable and the precision of the gene targeting is high, CRISPR/Cas9 has become the most attractive instrument for genome editing. Nevertheless, finding the ideal delivery method is challenging: AAVs are now the most employed vehicles for CRISPR/Cas9, as they possess several important benefits, including gene transferability to quiescent cells such as hepatocytes, absence of pathogenicity and low immunogenicity. Many AAV serotypes with different tropism have been identified, including AAV8 which has the greatest liver transduction efficiency, but they still raise some concerns about their cargo capacity and, above all, about their capacity to vehiculate the genome editing tool right in the target site [6].
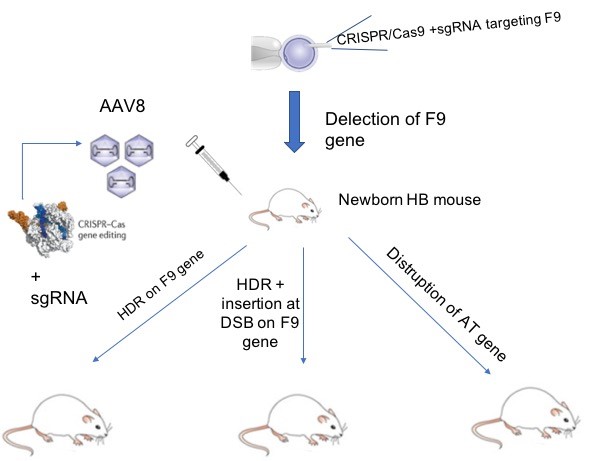
Figure 1: Generation of newborn Haemophilia B and phenotypic correction using CRISPR/Cas9.
In a paper written by a Japanese group, Ohmori and collaborators applied CRISPR/Cas9 –mediated genome editing using an AAV8 vector to treat haemophilia B in newborn mice [7]. Genome editing of newborns using AAV vector shows several important benefits, including avoiding regular replacement therapy beginning as early as 1-year-old: this means an improvement in the quality of life and significant reduction of costs as the pathology is eradicated even before childhood. Finally, using an AAV vector is more suitable in younger patients because of the lower frequency of anti-AAV neutralizing antibody [7]. The authors of this paper generated the disease in newborn mice and at the end they restored haemostasis using three different strategies (Fig.1). To start, haemophilia B was induced in newborn mice via deletion of F9 (therefore causing a reduction in factor IX activity) using CRISPR/Cas9. Interestingly, Ohmori T et al. successfully treated the disease not only targeting and correcting F9 gene, but also using an alternative strategy involving disruption of the antithrombin (AT) gene Serpinc1 by CRISPR/Cas9.
Later, in order to increase factor IX:C and cure the bleeding disorder, Ohmori and collaborators performed two different approaches in vivo. Both targeted F9 using an AAV8 vector to deliver genome editing tools in the liver and corrected haemophilia B using the two mayor pathways that can repair DSBs: the non-homologous end joining repair (NHEJ) and the homologous direct repair (HDR). The NHEJ pathway mediates the direct ligation of the broken ends, resulting in disruptive insertions or deletions (indels) at targeted locus. HDR is based on the homologous recombination that happens between a donor template of DNA and the sequences flanking the point of break on the target DNA, generating a new corrected form of the gene. Because of this, while NHEJ is an error prone mechanism, HDR is considered more accurate as a mechanism of repair. It mainly occurs in the S/G2 phase of the cell cycle when DNA is replicated, so the sister chromatid can work as donor template. HDR rarely occurs in post-mitotic adult tissues, such as skeletal muscle and the liver. In addition, HDR does not work well in neonates, because hepatocytes in neonates proliferate extensively [7].
To enhance precise genome editing, HDR is preferred and can be applied providing a donor sequence with homology arms on the target gene. Otherwise, an insertion mediated by NHEJ at DSB occurs. Several strategies can be applied in order to facilitate HDR, including the suppression of NHEJ using inhibitors or RNA interference. However, in Ohmori and collaborators’ paper, the expression of two adenovirus proteins (E1B55k and E4orf6) by AAV8 to increase HDR failed to increase factor IX in mice affected by haemophilia B. The authors demonstrated that phenotypic correction via HDR was less effective at increasing factor IX plasma levels compared with the AAV8-mediated insertion of a correct sequence without homologous arms at the DSB site. Interestingly, Ohmori and collaborators indicated that the most straightforward way to treat genetic diseases with genome editing is to enhance the insertion of the target sequence at DSB [7]. Finally, in order to correct the bleeding tendency of the disease, the researchers used an alternative strategy involving disruption of antithrombin gene (Serpinc1). Antithrombin is a serine proteinase inhibitor, which shows an important inhibitory activity of thrombin and other coagulation proteinases. Thrombin is a serine protease that converts soluble fibrinogen into insoluble strands of fibrin, which together with platelets form a clot over a wound site. Disruption of Serpinc1 resulted in a reduction of plasma AT activity, which improved fibrin formation after endothelial disruption in vivo.
CRISPR/Cas9 is definitely the most innovative technique for gene correction and seems to have a fine future ahead as far as it has been observed so far: easy production, low immunogenicity and a truly high rate of success. It has even overcome the use of zinc finger nucleases (ZFNs), whose potential and applications had been previously studied also in the treatment for haemophilia B [8]. Ohmori and his collaborators employ CRISPR/Cas9 not only to promote a “classical” correction of the gene, but also to inhibit the expression of AT, and this represents an alternative manner of use of this genome editing tool. On the other hand, even if CRISPR/Cas9 system enables efficient haemophilia B correction, one of the major hurdles to clinical application is the possibility of off-target effects which may lead to malignant transformations. Interestingly, the authors assessed 28 potential off-target sites and did not detect any mutations. However, off-target mutations should be carefully assessed before the clinical application in human therapy.
References
- Franchini M., Frattini F., Crestani S., Bonfanti C., Treatment of haemophilia B: focus on recombinant factor IX. Targets and Therapy 2013;7:33-38. doi: 10.2147/BTT.S31582
- Youjin S., Jun Y. The treatment of haemophilia A: from protein replacement to AAV-mediated gene therapy. Biotechnol Lett 2009;31:321–328. doi:10.1007/s10529-008-9869-0.
- Lheriteau E., Davidoff A. M., Nathwani A. C. Haemophilia gene therapy: Progress and challenges. Blood Reviews 2015;29:321–328. http://dx.doi.org/10.1016/j.blre.2015.03.002.
- Barrangou R. The roles of CRISPR–Cas systems in adaptive immunity and beyond. Current Opinion in Immunology 2015, 32:36–41. doi:10.1016/j.coi.2014.12.008.
- Jiang F., Doudna J. A. CRISPR–Cas9 Structures and Mechanisms. Rev. Biophys.2017,46:505-529. https://www.annualreviews.org/doi/abs/10.1146/annurev-biophys-062215-010822.
- Ohmori T., Mizukami H., Ozawa K., Sakata Y., Nishimura S., New approaches to gene and cell therapy for haemophilia. Journal of Thrombosis and Haemostasis 2015,13 (Suppl. 1): S133–S142. doi: 1111/jth.12926
- Ohmori T., Nagao Y., Mizukami H., Sakata A., Muramatsu S., Ozawa K., Tominaga S., Hanazono Y., Nishimura S., Nureki O., Sakata Y. CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B in mice. Scientific Reports 2017;7:4159. doi:10.1038/s41598-017-04625-5.
- Li H., Haurigot V., Doyon Y., Li T., Wong S. Y., Bhagwat A. S., Malani N., Anguela X. M., Sharma R., Ivanciu L., Murphy S. L., Finn J. D., Khazi F. R., Zhou S., Paschon D. E., Rebar E. J., Bushman F. D., Gregory P. D., Holmes M. C., High K. A. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 2011;475(7355):217-21. doi:10.1038/nature10177.

