Epigenetic modifications: building a synthetic propagation system in human cells
Minhee Park, Ahmad S. Khalil, and their collaborators, at the University of Boston, studied a synthetic method capable to regulate epigenetic modification in humans cells (HEK293FT)[1]. This method was based on the introduction of the modification m6A (N6-methyl-adenine) by the E.coli DNA adenine methyltransferase (Dam), a system that is widely diffused in bacterial species [2]. This could provide a simple model to study epigenetic interactions. The synthetic machinery involves three main modules: a synthetic initiator (made by ZF-Dam), a read-write module (made by DpnI-Dam ) and a synthetic reader module (made by DpnI-transcriptional factor (VP64)). The system was applied on two different gene reporter coding for GFP protein. The aim of this work was to create a synthetic system capable to introduce a methylation, maintain it through time and cell division, and finally affect gene expression. In order to build the simplest model possible, they chose to transplant a bacterial methylation system to avoid random cross-talk or interference with pre-existing network in the cells. Moreover, they build to kind of specific gene reporter to assess spatial propagation (clustered reporter) and temporal maintenance (interspersed reporter). The first had ZF harboring sequences preceding the GATC methylation motifs, while the second one has alternating ZF binding and GATC.
The first step of this work was to design a suitable initiator (SynI). The initiator had to be inducible and able to bind only specific regions. To achieve this goal they used a Dam library to find one with very impaired DNA binding system. The best Dam variant was found to be N132A. The initiator was then tested on the clustered reporter gene that showed an enrichment of the m6A. In order to make the initiator inducible they separated the ZF and Dam, binding them to two different transducing proteins, that can bind together only in presence of abscissic acid (ABA) [3]. Then they build an effector capable (formed by the fusion of DpnI and transcription factors) to read the methylated sequence and regulate reporter transcription. They did so by comparing two different signals one of a transcriptional promoter and one of a transcriptional repressor, both guided by the reader with the same transcription factors, directly guided by a ZF motifs.
Furthermore they tried to characterize the read-write module (SynRW). From the screening of the phenotype, as before they build a synRW library and tested it on the interspersed reporter. Comparing cells in active division not having the RW module against the one who had it, they managed to demonstrate that the presence of RW module is essential to maintain epigenetic memory through cells division, thanks to a positive feedback mechanism. And it can do so, even in absence of an external inductor signal (ABA). Moreover, the test was effective both in presence of a strong promoter (pUBC) and in the presence of weak promoter (pMinCMV). The best Dam variant for RW module was found to have an intermediate phenotype (R195A), between WT Dam (too much active) and Dam double mutant (not enough active).
Furthermore, they assessed the bidirectional propagation, placing a methylation with the CRISPR/Cas9, guided Dam right in the middle of the reporter, finding that the propagation is bidirectional as suggested by previous work [4]. The authors defined an optimum minimum distance between two GATC motifs that is 20 bp. They did so, comparing the methylation level among different reporters, with increasing distance between GATC motifs (20 bp, 50 bp, 212 bp, 800 bp). Finally, they proved, that in absence of the synR, in a system continually induced by ABA, the methylation’s degree is enhanced, so the synR can be used to modulate the degree of methylation in the cell, with a mechanism based on competitive inhibition.
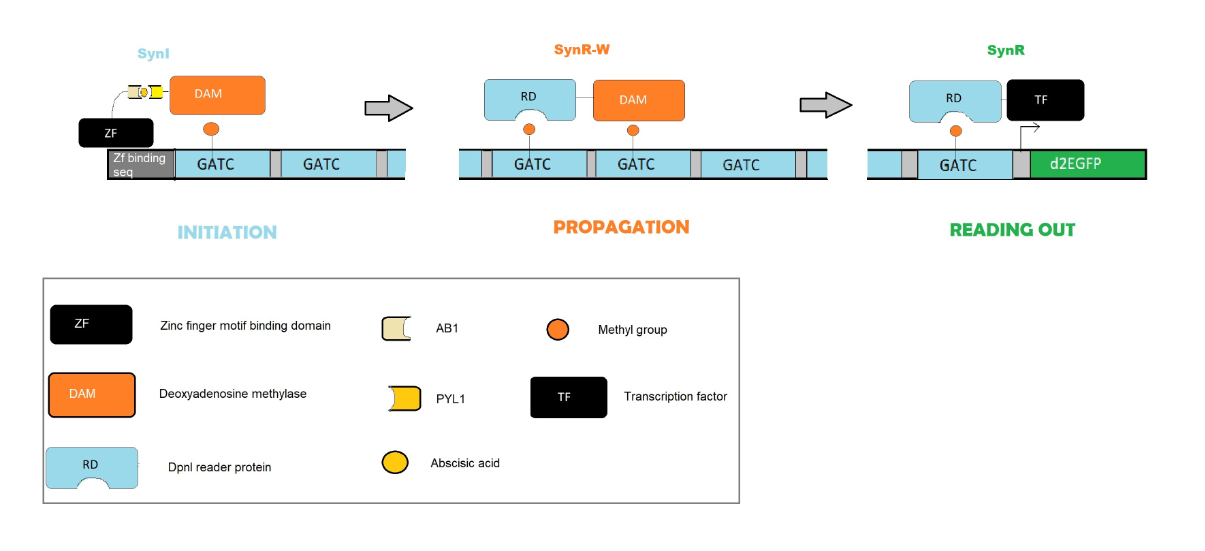
Fig 1.
This work shows that is quite easy to establish epigenetic modification using a synthetic regulation system that mimes natural systems of very simple organisms and proofs that what we learned so far about methylation can be applied in different organisms. However, these results do not add anything new to our knowledge about epigenetic systems. Moreover, it is very difficult to imagine the application of this technique in a commercial, clinical, or industrial environment, because the Dam can only methylate one DNA strand, not both and the article did not address the effectiveness of this system on human cells with WT sequences.
In our opinion it would be more interesting to try to develop a system that can modulate proper epigenetic systems of mammalian cells such as Cytosine methylation or histone modifications. Concerning the guiding system used in this work, it would be interesting to deepen the CRISPR/Cas9 models, also because one of the great limits of this method is the great amount of off-target sequences that, in our opinion, were not taken into account even if they represent a threat to the system efficiency.
References
- Park, M., Patel, N., Keung, A. J., & Khalil, A. S. (2019). Engineering Epigenetic Regulation Using Synthetic Read-Write Modules. Cell, 176(1–2), 227-238.e20.
- Gardner, K.E., Allis, C.D., and Strahl, B.D. (2011). Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409, 36–46.
- Liang, F.S., Ho, W.Q., and Crabtree, G.R. (2011). Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signal. 4, rs2.
- Hathaway, N.A., Bell, O., Hodges, C., Miller, E.L., Neel, D.S., and Crabtree, G.R. (2012). Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460
