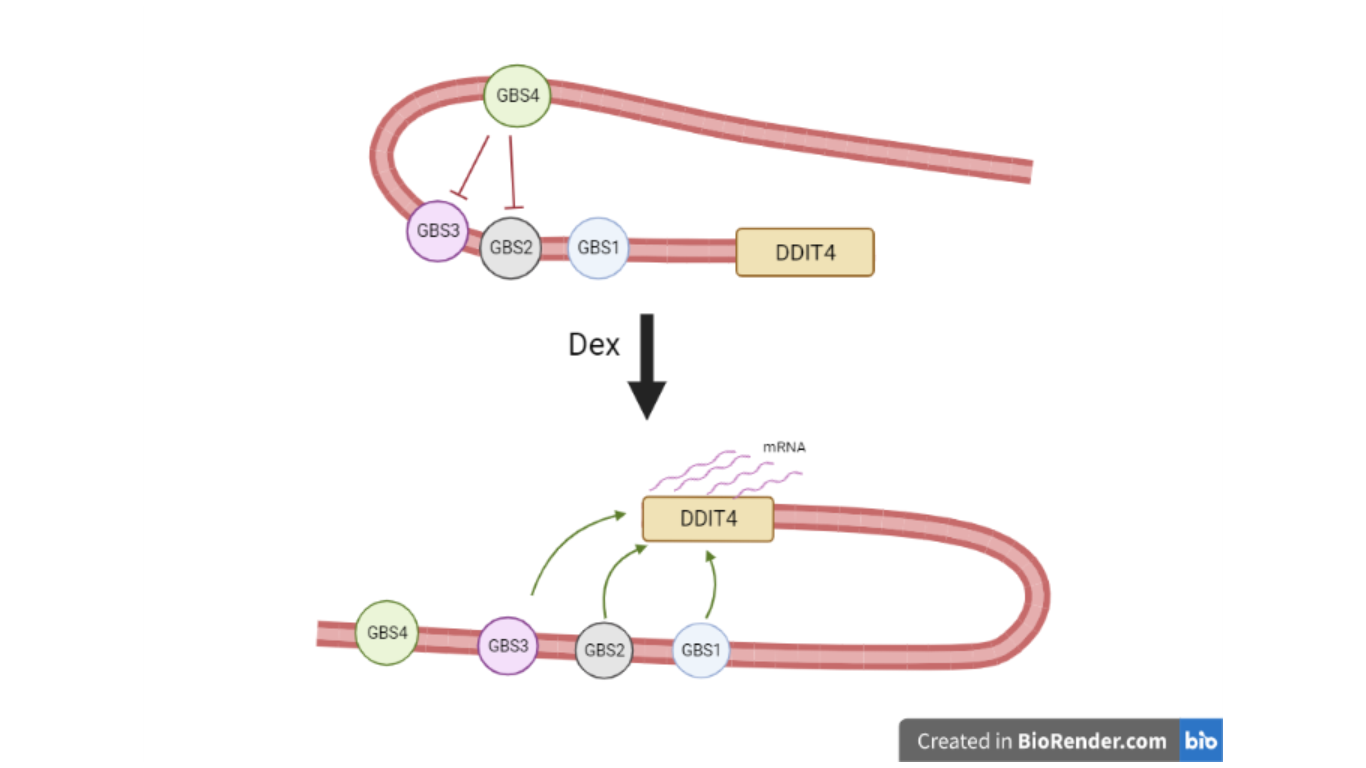
Mechanisms involved in Dex-induced transcription
Figure 1 – Dex induces a conformational change which blocks the inhibition mediated by GBS4
Abstract
Glucocorticoids are largely prescribed to treat various human diseases. They induce an extremely rapid and heterogeneous response. In a recent paper, Hoffman and his collaborators [1] tried to understand the mechanism behind this heterogeneity. They discovered how glucocorticoids induce a chromatin reorganization by forming new super enhancer regions which lead to the transcription of specific genes.
Discussion
Glucocorticoids are prescribed in medicine to treat a variety of human diseases such as cancer, asthma, and auto-immune disorders. Once inside cells, they bind to GRs (glucocorticoid receptors) located in the cytoplasm. This binding creates a complex which can go inside the nucleus and interact with DNA sequences called GBSs (GR binding sites).
The interaction with GBSs alters the expression of thousands of genes in the whole genome. Usually, GBSs have an active role in the transcription, and they are located in super enhancers (SEs), regions that contain more enhancers. [2,3] Glucocorticoid-mediated expression is highly heterogeneous. Hoffman et al. [1] led a study to identify the effects of Dexamethasone (Dex), a synthetic glucocorticoid, on human breast cancer cells. The authors based their studies on two cultures of breast cancer cells T47D, A1 and A2, treating them with ethanol as control (Veh) and dexamethasone in ethanol, respectively.
The first step was identifying genomic changes in acetylation of lysine 27 on histone H3 (H3K27ac) as a consequence of Dex treatment. Such acetylation is a clear indication of the chromatin de-condensation which makes it more accessible to polymerase to start the gene transcription. By performing a H3K27ac chromatin immunoprecipitation sequencing (CHIP-seq) following the Dex treatment, the authors observed a reorganization of chromatin by acetylation; some poorly acetylated regions increased their level of acetylation, whilst others lost it.
Due to this reorganization, they could observe the activation and inactivation of about one hundred of SEs.
To understand the mechanisms behind the heterogeneity of the transcription in glucocorticoid-activated genes, the SE of the DNA Damage Inducible Transcript 4 oncogene (DDIT4) was taken as a representative of Dex-induced SE. By GR CHIP-seq, they could define the structure of the DDIT4 SE, which is composed by four GBSs located respectively at 19, 20, 25 and 30 kb upstream of the DDIT4 transcription start site (TSS) of DDIT4.
In the first part of the study they have verified the transcriptional heterogeneity of DDIT4 using a single cell RNA-seq (scRNA-seq) and subsequently by using a single molecule Fluorescence in Situ Hybridization (smFISH). After the confirmation of the suitability of DDIT4 as a model for the study, the authors continued by investigating the function of GBSs located in DDIT4 SE.
Initially, the authors cloned every GBS of the DDIT4 SE to a plasmide vector, upstream or downstream the luciferase gene, obtaining interesting results. In fact, it seemed that every GBS, taken individually, could activate the transcription of the luciferase gene.
So, to see if GBSs act in a different way in a SE region, the authors used the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technique to create knock out lines for each GBS, to analyze the native and mature mRNA of DDIT4 by Real-time PCR. From that experiment, they could show that the removal of GBS1 and GBS3 caused a slowdown in mRNA production while the removal of GBS2 caused a dramatic reduction in DDIT4 mRNA production. In contrast, removing the GBS4 from the SE gave the most unexpected results: in the GBS4 KO line, there was an exponential increase in DDIT4 production. These results made it possible to attribute to the GBS1, GBS2 and GBS3 the function of an enhancer whilst GBS4 acted as a silencer of the DDIT4 gene. It is extremely interesting to notice how the GBS4, when isolated, acts as an enhancer in the transcriptional process, while when it cooperates with other GBSs, it is identifiable as an inhibitor.
The second part of the study aimed at delineating the dynamics of interactions between GBSs regions. Authors proposed the involvement of two mechanisms of interaction. They analyzed the presence of binding sites of transcription factors and the formation of chromatin loops to allow interaction between distant DNA regions. Authors used the JASPAR 2020 software [4], to predict transcription factors (TFs) specific for the GBSs which could characterize their activity. The software found only two transcription factors specific for the GBS4 which were E2F1 and E2F2; the others were common for every GBS. Anyway, using a ChIP-PCR, they saw that these two TFs could bind the GBS2 and the TSS instead of the GBS4. Thanks to these results, it was possible to affirm that activity of the GBSs isn’t due to specific transcription factors.
To analyze the presence of chromatin loops they investigated on the sites involved in the formation of these loops such as CCCTC-Binding Factor (CTCF) and the cohesin subunits RAD21 and SMC3. The results obtained by CHIP-qPCR showed an enrichment of RAD21 and SMC3 at the TSS of DDIT4 and on each GBSs, thus suggesting the involvement of chromatin loops in the interactions between GBSs and the TSS.
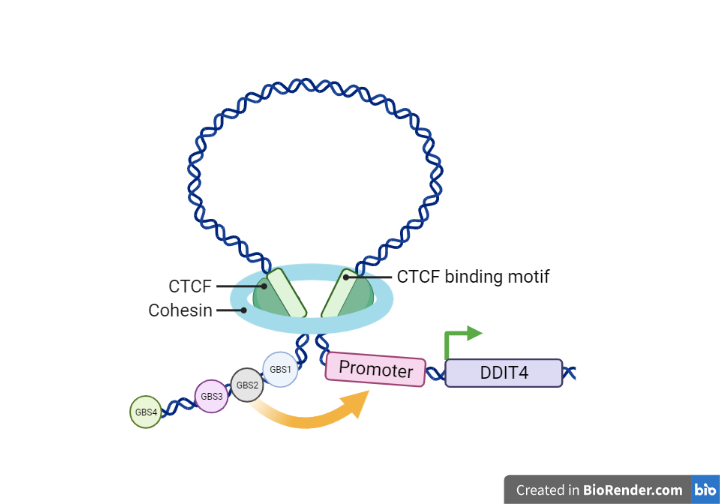
Figure 2 – Representation of the chromatin loop
To further confirm these results, they performed a 4C-seq using the TSS viewpoint and it was observed that, following Dex treatment, the TSS region interacts exclusively with GBS1, GBS2 and GBS3. Interestingly, there are no interactions between GBS4 and the TSS.
These results further suggest the inhibitory function of GBS4 and the activatory role of GBS1, GBS2, and GBS3, which are probably due to the formation of chromatin loops.
Furthermore, to investigate these interactions between GBSs, the same technique has been performed on individual KO lines. The results showed that the removal of a single GBS leads to an almost complete reduction of the interactions of the TSS with the remaining GBSs.
Consequently, the presence of all its components is necessary for the SE to act properly. Based on these studies, they hypothesized that the heterogeneity of DDIT4 expression is caused by the stochasticity of the interactions mediated by chromatin loops and by the presence of the different transcription factors at the level of GBS.
Conclusions
We found this article appealing because it offers a new perspective on the regulation of the transcriptional process, demonstrating how the latter is a spectrum of complex mechanisms which are almost entirely undiscovered, such as the level of regulation based on the interaction between regions distant from each other. Having full knowledge of these mechanisms, it will be possible to use glucocorticoids, minimizing negative side effects.
However, this study was performed in vitro. In order to confirm the results obtained by Hoffman et al. are needed further in vivo studies.
References
- J.A. Hoffman, K.W. Trotter, C.R. Day, J.M. Ward, K. Inoue, J. Rodriguez, T.K. Archer, Multimodal regulatory elements within a hormone-specific super enhancer control a heterogeneous transcriptional response, Molecular Cell. 82 (2022) 803-815.e5. https://doi.org/10.1016/j.molcel.2021.12.035.
- D. Hnisz, B.J. Abraham, T.I. Lee, A. Lau, V. Saint-André, A.A. Sigova, H.A. Hoke, R.A. Young, XSuper-enhancers in the control of cell identity and disease, Cell. 155 (2013) 934. https://doi.org/10.1016/j.cell.2013.09.053.
- W.A. Whyte, D.A. Orlando, D. Hnisz, B.J. Abraham, C.Y. Lin, M.H. Kagey, P.B. Rahl, T.I. Lee, R.A. Young, Master transcription factors and mediator establish super-enhancers at key cell identity genes, Cell. 153 (2013) 307–319. https://doi.org/10.1016/j.cell.2013.03.035.
- Fornes, J.A. Castro-Mondragon, A. Khan, R. van der Lee, X. Zhang, P.A. Richmond, B.P. Modi, S. Correard, M. Gheorghe, D. Baranašić, W. Santana-Garcia, G. Tan, J. Chèneby, B. Ballester, F. Parcy, A. Sandelin, B. Lenhard, W.W. Wasserman, A. Mathelier, JASPAR 2020: Update of the open-Access database of transcription factor binding profiles, Nucleic Acids Research. 48 (2020) D87–D92. https://doi.org/10.1093/nar/gkz1001.