Molecular aspects of the interaction between host epigenome, gut microbiota and diet
Abstract
Due to the spread of a lifestyle based on the consumption of diets with a high percentage of fat, the rate of obesity has grown in last years. Obesity carries a rate of chronic inflammation and gastrointestinal diseases such as colon cancer [1,2]. The transplantation of obesogenic diet-conditioned microbiota into germ-free mice, combined with a fat rich diet, recapitulates the features of the long-term diet regimen. These findings suggest that the gut microbiome, under specific dietary exposures, stimulates the reprogramming of the enhancer landscape in the colon, with downstream effects on transcription factors.
The symbiotic relationship between the guest and the microbiota has a fundamental importance, bacteria are involved in digestion, immunological development and epithelial homeostasis of the host [3]. Therefore, studying how the host’s epigenome responds to the diet can give an overview. This is what Yufeng Qin and his team, in 2018, tried to bring to light in their study [1]. A mouse model has been used to define the correlation between the host’s epigenome, microbiota and diet. Male and female mice were taken and divided into two different diet regimens: one fed with a low-fat diet (LFD) and the other with a high-fat diet (HFD) and monitored over time. As a result, were found different families of bacteria and weight variation in a sex-specific manner. Microbiota of diet-induced obesity alters the acetylation of Lys27 on histone H3 (H3K27ac) and the monomethylation of Lys4 (H3K4me1).
To see whether these genes were expressed differently in obese mice and whether could resemble the changes in the human rectal cancer scenario, they compared the 10 most misexpressed genes in their study with Oncomine data sets. The major changes, caused by obesity in the mice, are surprisingly close to the changes that arise in colon adenocarcinoma. In H3K27ac enriched loci, one of the most enriched motifs corresponds to HNF4α, a nuclear receptor, which is reprogrammed by obesity.
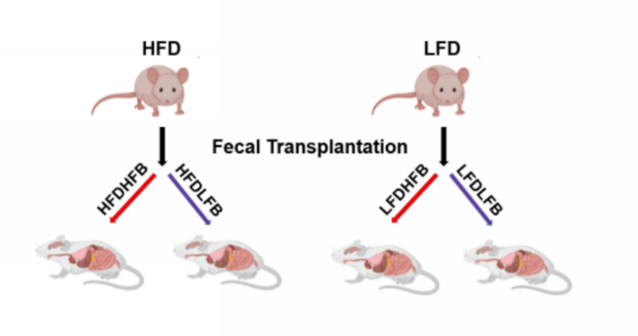
Figure 1. Bacterial transplantation.
Afterward samples of bacteria, belonging to the microbiome of male donor mice fed with LFD or HFD, were taken and introduced in 6-week male and female wild-type mice fed with LFD or HFD (Figure 1). Interestingly, male mice fed with the obesogenic diet (HFD) and those who had received a bacterial transfer from obese animals (HFDHFB) gained more weight compared to those who received bacteria from the controls (HFDLFB). ChIP-seq has been carried out in HFB to characterize the relationship between bacterial status, diet on enhancers. Genes associated with differentially enriched loci in mice fed with HFD, showed similar profiles to the gene sets expressed in tumors of the digestive system and gastrointestinal neoplasms.
Since it was understood that HFD-HFB had reshaped the epigenome similarly to colon cancer-related landscape, the authors compared top ten misexpressed genes related to colorectal cancer from Oncomine. The results indicated that the combination of HFD-HFB may induce a gene expression profile that has a partial similarity to that observed in human colorectal cancer. Using GSEA has been seen that the genes regulated by the nuclear receptor HNF4α were enriched in the downregulated genes in the HFD-HFB group compared to the HFD-LFB group. HNF4α has been reported to recruit co-repressors to inhibit integral gene expression at lipid homeostasis in the liver and maintaining intestinal homeostasis in response to microbiota changes.
To understand the regulatory role of HNF4α, were carried out ChIP-seq analysis in the HFD-HFB and HFD-LFB groups, to determine the genes that are downregulated from HNF4α into HFD-HFB. In HFD-HFB is observed an increased of HNF4α binding at the Fmo1/2 loci related to the reduction of these genes expression. FMO1 and FMO2 play an important role in iron metabolism and regulate the formation of reactive oxygen species. Genetic deletion of Hnf4α in mice reduces the binding to the Fmo 1/2 loci and increases their expression levels [4]. Similarly, SCD1, a key enzyme for the synthesis of fatty acids, is downregulated in the liver under fasting conditions, which increase the level of HNF4α, correlating with a reduced expression of SCD1 [5].
Resultantly is evident that the intestinal microbiota participates in the regulation of gene expression through DNA methylation in intestinal epithelial cells. The gut microbiota changed the state of the open chromatin in the intestinal intraepithelial, producing dysbiosis linked to the pathophysiology of obesity. The obesogenic diet resulted in a decrease in “beneficial” flora causing a very different microbial profiles, between lean and obese animals, associated with the onset of chronic inflammation, a factor that predisposes to cancer [6].
This study can highlight that there are potential interactions between the host diet and the microbiome and this affects the epigenome of the host. Furthermore, host-microbiota interactions have potential relevance for obesity and obesity-related diseases.
References
- Yufeng Quin, John D Roberts, Sara A Grimm, Fred B Lih, Leesa J Deterding, Ruifang Li, Kaliopi Chrysovergis, Paul A Wade. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome biology, 2018, doi: 10.1186/s13059-018-1389-1
- Kopelman PG. Obesity as a medical problem. Nature, doi: 10.1038/35007508
- Winer DA, Luck H, Tsai S, Winer S. The intestinal immune system in obesity and insulin resistance. Cell Metab, 2016, doi: 10.1016/j.cmet.2016.01.003
- Matsuo S, Shunsuke Matsuo, Masayuki Ogawa, Martina U Muckenthaler, Yumiko Mizui, Shota Sasaki, Takafumi Fujimura, Masayuki Takizawa, Nagayuki Ariga, Hiroaki Ozaki, Masakiyo Sakaguchi, Frank J Gonzalez, Yusuke Inoue. Hepatocyte nuclear factor 4alpha controls iron metabolism and regulates Transferrin receptor 2 in mouse liver, J Biol Chem, 2015 doi:1074/jbc.M115.694414
- Xie X, Liao H, Dang H, Pang W, Guan Y, Wang X, Shyy JY, Zhu Y, Sladek FM. Down-regulation of hepatic HNF4alpha gene expression during hyperinsulinemia via SREBPs. Mol Endocrinol, 2009; doi: 10.1210/me.2007-0531 (6)
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. 2011; doi: 10.1016/j.cell.2011.02.013