scRNA-seq reveals a high reproducibility of the brain’s organoids: a valuable organoid model for studying human neurological diseases?
Abstract
Recently, scRNA-seq analysis of individual brain organoids has shown that an organoid model of the dorsal forebrain consists of a cellular diversity typical of the human cerebral cortex. This level of in vitro reproducibility allows to study the basic principles of the human brain development and it is also fundamental because it establishes a precious organoid model capable of experimental investigation on developmental anomalies associated with human neurological disease.
It is known that the main limit of the human brain organoids is that they are plagued by high organoid-to-organoid variability. Consequently, it was uncertain whether the brain development processes could be faithfully modeled in vitro. However, in 2019, some researchers, mostly from Harvard University (1), have tried to clear up this doubt. As a result, the researchers found that among all the 21 brain organoids they generated, about 95% of the organoids were made up of different cell types with a degree of organoid-to-organoid variability comparable to that of individual endogenous brains, demonstrateing that the brain development processes can carefully take place even outside the embryo context.
Previous studies have shown that a wide variety of cell types, characteristic of different brain regions, can be generated in vitro from pluripotent stem cells. However, few studies have estimated the cellular composition of the generated organoids, not allowing to delineate the degree of reproducibility between them. For this reason, the researchers began their study by generating, through 4 distinct protocols, the brain organoids, deriving all from the same human iPS cell lines (PGP1). The dorsally patterned organoid model was the one with the best morphological characteristics on which an immunohistochemical analysis was carried out. The dorsal forebrain protocol was applied on five distinct cell lines (PGP1, HUES66, GM08330, Mito 210 and 11a) and 100% of organoids expressed dorsal forebrain progenitor markers (PAX6, EMX1) at one, three and six months of growth. On the basis of these
promising results, single-cell RNA sequencing (scRNA-seq) was performed on the dorsally patterned organoid model.
scRNA-seq analysis provides deeper insight to the multi-tiered complexity of different cells within the same tissue type. This analysis was applied on nine organoids at 3 months of growth, derived from 2 different stem cell lines (PGP1, HUES66) and was the fulcrum of this study because it characterized eleven main transcriptionally distinct cell types across both lines, appropriate for the cerebral cortex. Subsequently, the authors aligned and grouped the cells of all organoids showing that there was a high degree of reproducibility in the cellular composition between the different organoids. These data suggest not only that these organoids are composed of different types of cortical cells but that their identity and cellular variability can be reproducible.
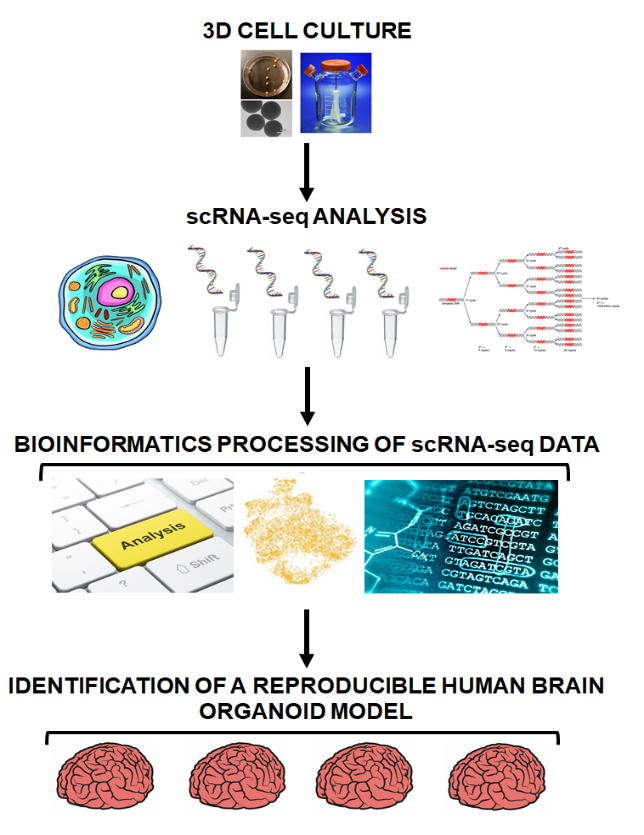
Fig. 1 Scheme of the procedure followed for the analysis of the reproducibility and plausibility of the model.
It is well known that cellular diversity of the cerebral cortex develops in a specific temporal sequence. Based on this concept, the authors performed a new scRNA-seq analysis of twelve organoids from 3 different human iPS cell lines (PGP1, 11a and GM08330) after six months of development, which confirmed that organoid to organoid variability remained extremely low even at that stage of culture. In addition, to demonstrate the change in cellular composition over time during the development process, the authors compared the cellular composition of two organoids derived from the same line (PGP1) but grown for different times (three months vs six months). Also confirmed by immunohistochemistry, the six-month cultures contained substantial numbers of astrocytes, cells that are generated not initially during development, showing that the development process of these organoids respects endogenous temporal sequences.
It is however possible that the same cellular composition between the different organoids could be generated through distinct developmental Fig.1 trajectories. To answer this question, a pseudotime developmental trajectory between all cell lines was calculated.
Most cell-state transitions, whether in development, reprogramming, or disease, are characterized by cascades of gene expression changes. Pseudotime temporal ordering applies machine learning to scRNA-seq data to order cells along a reconstructed ‘trajectory’ of differentiation or other type of biological transition. This approach is based on the fact that the cells during their differentiation path make strong decisions with the aim of specializing in a specific cell type and these decisions certainly involve changes in the gene expression of the cells. This approach was applied to the study conducted and showed that each organoid, regardless of the group considered, is distributed similarly along the development trajectory, also highlighting that this order of temporal development is approximate to that of human development.
Next, a published scRNA-seq dataset of human fetal cerebral cortex was used to train a random forest classifier. The random forest classifier (a virtual machine), after learning brain endogenous data, was able to assign organoid cells to the human cortex cell categories. The results showed high correspondence between organoid cells at both three and six months and fetal cerebral cortex cell class, indicating a correlation in their transcriptional profiles.
In conclusion, this research demonstrates, by performing scRNA-seq analysis, that it is possible to obtain, in an experimentally reproducible way, a human brain organoid model able to generate several cell types of the human cerebral cortex. The discovery of an organoid model that faithfully reproduces the embryogenesis of the neuronal tissues will be used in the experimental investigation of developmental abnormalities associated with human neurological disease.
However, the results of this research article come almost completely from bioinformatic analyzes, therefore it may be necessary to perform further histological studies to confirm the reliability of the brain organoid model. Moreover, the cells form a specific structure and 3D cellular organization during the embryogenesis of the nervous tissue, therefore it would be useful to investigate whether these features are maintained in the same way in organoids. At last, the experimental development of human brain models in vitro has raised some ethical issues that are still debated among scientists. In some cerebral organoids, neural connections and electrical activity can be measured with some techniques that are already available (the Perturbational Complexity Index used for brain-injured non-communicating patients). In the future, if brain organoids will show a glimpse of sensibility, an ethical discussion on their use in clinical research and practice will be necessary.
References
- Velasco, S. et al. “Individual brain organoids reproducibly form cell diversity of the human cerebral cortex”. Nature, 570(7762), 523–527 (2019).
- Quadrato, G., Brown, J., & Arlotta, P. “The promises and challenges of human brain organoids as models of neuropsychiatric disease”. Nature Medicine, 22(11), 1220–1228 (2016).
- Quadrato, G. et al. “Cell diversity and network dynamics in photosensitive human brain organoids”. Nature, 545(7652), 48–53 (2017).
- Hwang, B. et al. “Single-cell RNA sequencing technologies and bioinformatics pipelines”. Exp Mol Med 50, 96 (2018).
- Qiu, X. et al. “Reversed graph embedding resolves complex single-cell trajectories”. Methods 14, 979–982 (2017).
- Rigamonti, A. et al. “Large-scale production of mature neurons from human pluripotent stem cells in a three-dimensional suspension culture system”. Stem Cell Reports, 6(6), 993–1008 (2016).
- Lavazza, A., & Massimini, M. “Cerebral organoids: Ethical issues and consciousness assessment”. Journal of Medical Ethics, 44(9), 606–610 (2018).

