The importance of non-coding variants in neuropsychiatric disorders
Abstract
An integrative analysis of chromatin interactions, open chromatin regions and transcriptomes has been performed in four neural cell types to link neuropsychiatric disorder risk variants to target genes.
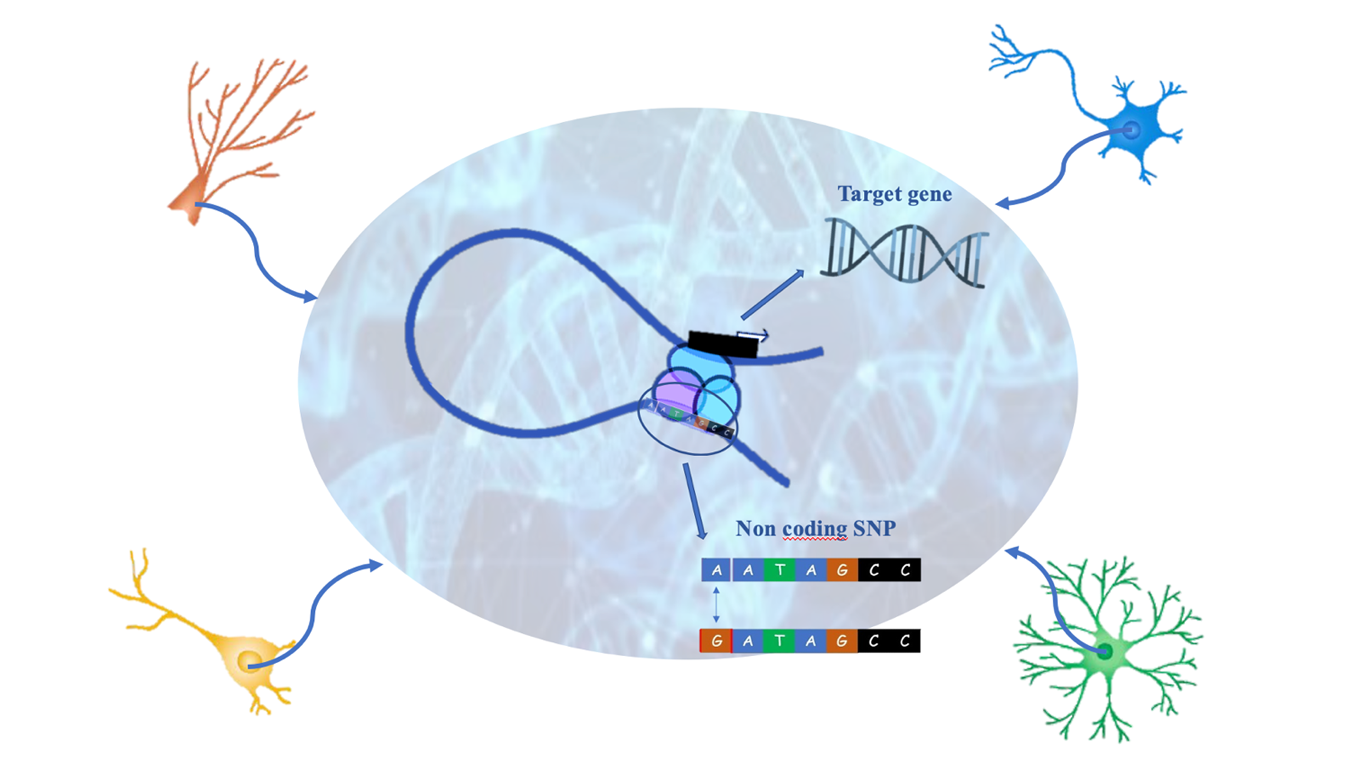
Many genetic variants associated with human traits and diseases are located in putative gene regulatory elements, and these can contribute to human diseases by modulating and altering gene expression. The role of these noncoding variants is not easy to study, because of the lack of annotation in the physiologically relevant cell types and because of the limits of the nearest gene model (neighboring genes assigned as risk loci for noncoding variants). Indeed, there are many experimental evidences that regulatory elements often interact with their target genes over long distances [1]. For these reasons the three-dimensional chromatin structure is very important in gene regulation and diseases, also in the human brain during neurogenesis and for the onset of neuropsychiatric disorders [2, 3]. Moreover, noncoding variants contribute to diseases in an intricate and cell-type specific manner [4], and especially in the field of neuroscience there is a lack of annotations.
To identify causal variants, their target genes, and subsequently deciphering their functions Song et al. [5] had mapped the landscape of epigenomic regulation in well-characterized, physiologically relevant neural cell type: iPSC-induced Excitatory neurons, iPSC-derived hippocampal DG-like neurons, iPSC-induced lower motor neurons and primary astrocytes isolated from 19-week-old male fetal brain samples. They used an integrative analysis of chromatin interactions, open chromatin regions and transcriptomes for each cell type [Figure 1].
First, Song et al. have characterized the epigenomic landscape of long-range chromatin interactions. They discovered that the majority of interactions between promoters and PIRs (Promoter Interacting Regions) are included in TADs (Topologically associated domains) and 40% of them are cell-specific: this is relevant because they have an important role in providing the main differences between cell types. Moreover, each promoter interacts on average with 7-8 PIRs and these PIRs are often enriched in chromatin-active regions resulting fundamental in transcription activation. Promoters interacting with enhancers show a bigger transcription level and its amount is proportionally related to the number of PIRs per promoter.
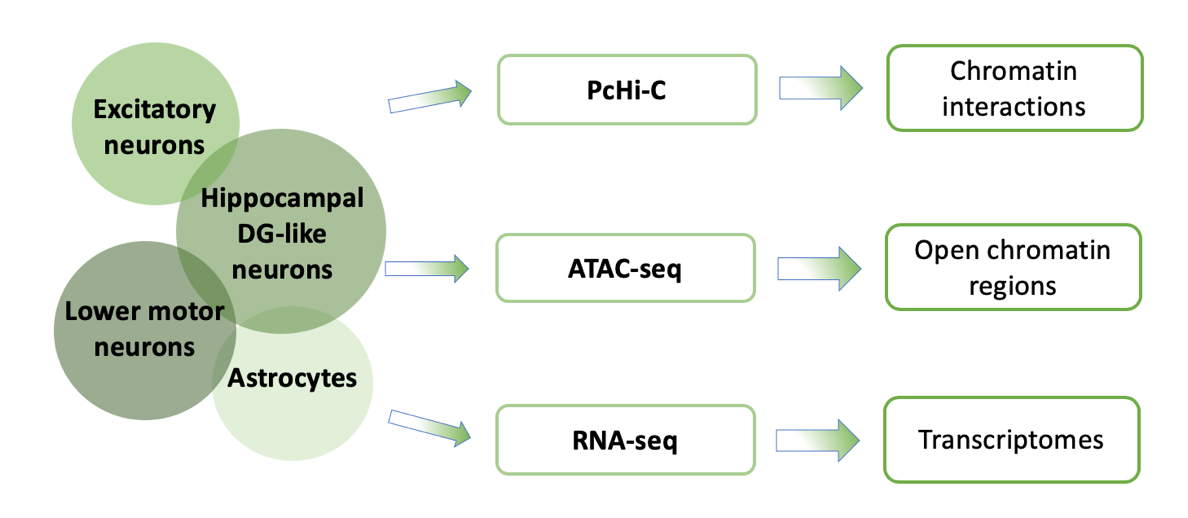
Figure 1: Integrative analysis of chromatin interactions using pcHi-C, open chromatin regions using ATAC-seq and transcriptomes using RNA-seq in each cell type.
Using Chicago score, the researchers were able to demonstrate that PIRs-promoter interactions are specific in the four cell types analyzed, and this has got an important role in underline functional differences between them, as demonstrate by the GO (Gene Ontology) enrichment analysis for genes involved in these specific interactions. In PIRs there are also cell-specific motifs of TF, which provide information in agreement with the GO.
To identify the targets of these PIRs, Song et al. considered the VISTA Enhancer Browser [6], a browser containing sequences validated as enhancers. They compared them with data from their experiments and found that about one third of their interactions involves sequences from VISTA, and many of them were further annotated as neural enhancers. After that, they performed validation of PIRs using CRISPR interference technique to silence two PIRs sequences of CDK5RAP3 promoter, called region 1 and region 2.
It is known that lots of PIRs are involved in many neuropsychiatric disorders [7]; starting from GWAS Catalog [8], which contains SNPs related to these illnesses, researchers found that 70% of them are in non-coding regions. Moreover, lots of mutations specific for some diseases are enriched in some cell-types, for example schizophrenia SNPs (SCZ SNPs) are spread around the four typologies of SNC-cells, but unipolar depression SNPs (UD SNPs) are enriched only in excitatory and hippocampal neurons, so we can think that they are much involved in this disorder. Also in this case, they performed a validation of PIRs containing disease associated SNPs: for example SCZ SNPs interacting with DRD2 promoter which activate the transcription of DRD2 (a gene associated with anxiety and depression), have been silenced and the result was an important downregulation of DRD2.
Therefore Song et al. understood that SNPs in non-coding regions are very important because they modulate chromatin architecture and, as a result, gene expression, which in some cases can also cause disabling psychiatric disorders. This is also the main development that this work has delivered in the context of neuroscience studies, because GWAS experiments were usually based on SNPs in coding regions, since their effects are easier to establish. So, having improved this vision with non-coding SNPs and their related gene targets, which often underlie disabling illnesses, represents a great result. Moreover, they have discovered that these non-coding variants depends on where they are located, because many variants in PIRs are more enriched in some cell-types and less in others.
On the other hand, in the future it will be necessary to improve these epigenomic annotations based on PIRs, also including single cell-sequencing in order to have a deeper insight into complex disease biology. Regarding models, derived-patience cells could be useful for this purpose; alternatively, it will be suitable inducing specific mutations to obtain compromised interactions seen in diseased. Furthermore, the fine-tuning of organoids can give the same results but with a lower cost.
References
- Mumbach, M. R. et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nature Genetics 49, 1602–1612 (2017).
- de la Torre-Ubieta, L. et al. The dynamic landscape of open chromatin during human cortical neurogenesis. Cell 172, 289–304 e218 (2018).
- Won, H. et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016).
- Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014).
- Song, M. et al. Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nature Genetics 51, 1252-1262 (2019)
- Visel, A., Minovitsky, S., Dubchak, I. & Pennacchio, L. A. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–92 (2007).
- Roadmap Epigenomics, C. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
- Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).