A new weapon to eradicate malaria
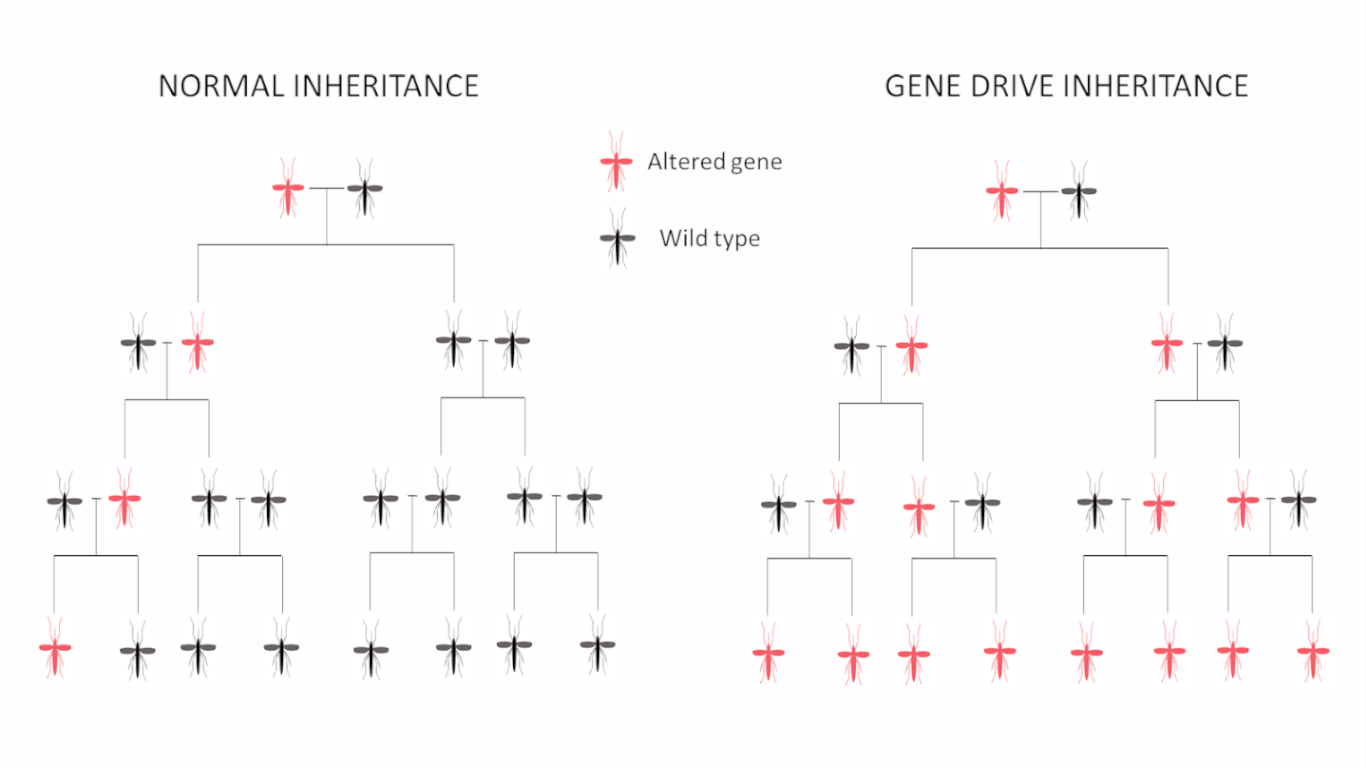
A CRISPR–Cas9 gene drive construct was used to target specific genes involved in female fertility in Anopheles gambiae, the human malaria vector. In particular, the intron 4–exon 5 boundary of gene doublesex was disrupted, generating sterile females and causing population collapse in caged mosquitoes within few generations.
Malaria is a potentially deadly infectious disease caused by a microorganism of the genus Plasmodium, which is transmitted to people exclusively through the bite of an Anopheles mosquito. Malaria represents a huge global health problem and is the main cause of mortality in many countries. The most recent data are eloquent: according to WHO world malaria report 2018, in 2017 about 219 million of cases of malaria have been identified worldwide, with a trend that does not seem to show a reduction of cases over time. For 92% of cases (more than 200 million) malaria cases were recorded in Africa, followed by a 5% of cases in the South-East Asia Region and a 2% of cases in the Eastern Mediterranean Region. In 2017, about 435000 deaths occurred worldwide, in which 61% of them were children under 5 years, that represents the main susceptible group potentially affected by malaria.
With the introduction of the new technologies for genome editing, Kyrou et al. [1 ] at the Department of Life Sciences of Imperial College London, developed a strategy in order to cause a complete population suppression in caged Anopheles gambiae mosquitoes, the main malaria vector. In particular, a CRISPR-Cas9 complex has been used in a gene drive construct to target specific female fertility genes of A. gambiae. This approach allows to suppress the reproductive capability of the mosquitoes, decreasing the number of fertile females in order to cause the population collapse.
In A. gambiae sex differentiation is controlled by an alternative splicing of the doublesex (dsx) gene. This gene is composed by seven exons and undergoes an alternative splicing to produce the male and female transcripts AgdsxM and AgdsxF: the latter differs for a highly conserved exon (exon 5) which is absent in AgdsxM transcript. The boundary between intron 4 and exon 5 was used as target for a gene drive test. In particular, it was disrupted so that the functional AgdsxF could not be formed while AgdsxM remained unvaried. A single-guide RNA (sgRNA) and Cas9 were used to recognize and cleave, respectively, the target sequence. A homology-direct repair (HDR) is able to insert a sequence carrying eGFP gene at the target site. Heterozygous individuals were crossed to generate wild type, heterozygous and homozygous for the dsxF– allele according to Mendelian ratio 1:2:1. dsxF+/+ and dsxF+/− individuals showed normal development and wild-type morphological characteristics, whereas 50% dsxF−/− are male which present a wild-type phenotype and the other 50% are female showing male specific morphological characteristics, such as plumose antennae and unstructured testes. dsxF-/- females were unable to bite mice to take blood and could not produce any eggs. This suggests that dsx is involved in sex differentiation pathway of A. gambiae and may be nominated as a target for a gene drive experiment designed for population suppression.
For this experiment it was used a drive construct, CRISPRh cassette, that contains the transcription unit of a Cas9 endonuclease, a RFP gene and the gRNA, targeting dsxF. The cassette was inserted at the target locus. During meiosis the Cas9/gRNA complex is able to cleave the wild-type allele at the target locus and the construct previously inserted is duplicated on the wild-type allele via HDR disrupting exon 5, bypassing Mendelian inheritance laws.
Crossing wild-type individuals with heterozygous dsxFCRISPRh/+ female or male mosquitoes, dsxFCRISPRh showed a high transmission to the progeny. Heterozygous dsxFCRISPRh/+ males had a similar larval progeny than wild-type mosquitoes, while heterozygous dsxFCRISPRh/+ female showed a fecundity rate lower than wild-type mosquitoes.
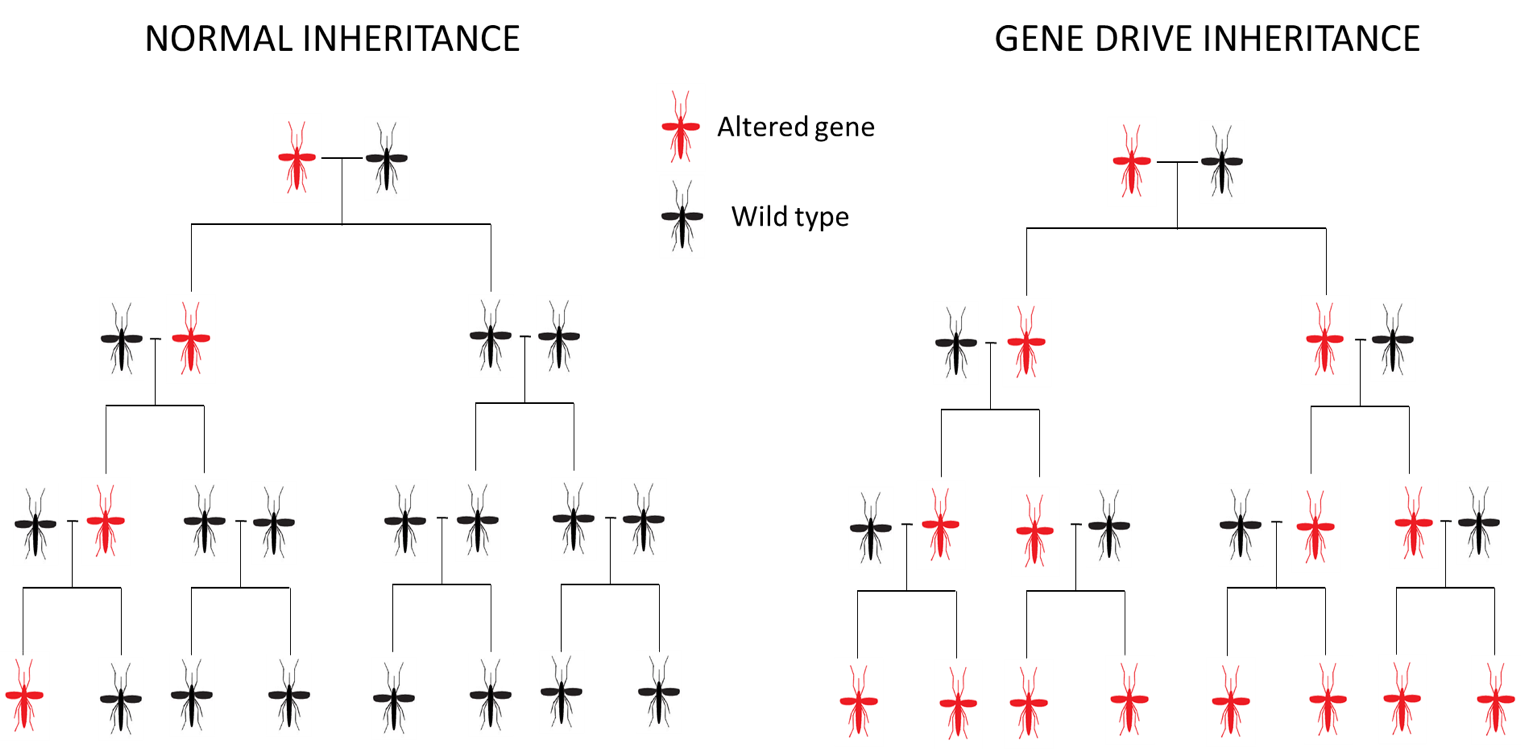
To evaluate the transmission frequency of the dsxFCRISPRh allele within a population, wild-type and heterozygous dsxFCRISPRh/+ mosquitoes were crossed into two replicate cages. dsxFCRISPRh allele reached 100% frequency in both cages at generations 11 and 7 and at the subsequent generation female mosquitoes were unable to produce any eggs and population collapsed.
The authors also evaluated the presence of mutations at the target site within the first 5 generations. The variants identified had not shown positive selection in the subsequent generations because of the high conservation of exon 5 in A. gambiae. Only a single SNP within the drive target site was detected in sub-Saharan African mosquitoes, nevertheless Cas9, guided by gRNA, was able to cleave efficiently the sequence containing the SNP.
It would be also necessary to evaluate dsx spread in native conditions, rather than laboratory environment. Indeed, mosquitoes can be susceptible to various environmental stressors, which may affect population fitness. A mathematic deterministic model shows that a reduction of fitness until 40% allows to gene drive to reach 100% frequency which results in population suppression.
Forasmuch as all the insects share the same dsx pathways in sex determination, a gene drive-based-technique could be applied to different insect pests (zika, dengue, yellow fever vectors). However, the specific introns and exons involved in the pathways of each insect family should be known to apply this gene editing approach.
In this study, researchers evaluated the onset of mutations at the target site in generation 2, 3, 4, and 5, which could give rise to functional variants or nuclease resistant. However, we cannot rule out the possibility that a mutation could arise and could be positively selected to restore the normal function of the gene and be spread to the offspring in the subsequent generations. On the other hand, malaria kills about 1000 individuals every day, so it is necessary to increase this research-line in order to guarantee the highest and quickest diffusion of the gene within the mosquito population.
References
- Kyrou et al. (2018) Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton A, Nolan T, Crisanti A. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiaemosquitoes. Nature Biotechnology. 2018; 36:1062–1066.
- Hammond, A. et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016).
- Scali, C., Catteruccia, F., Li, Q. & Crisanti, A. Identification of sex-specific transcripts of the Anopheles gambiae doublesex gene. J. Exp. Biol. 208, 3701–3709 (2005).
- The Anopheles gambiae 1000 Genomes Consortium. Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552, 96–100 (2017).
