Single-cell ChIP seq reveals cell subpopulations defined by chromatin state
Abstract
The application of recent technologies like microfluidics, DNA barcoding and next-generation sequencing has led to the chromatin profiling at single-cell resolution, which allows to distinguish the identity of each single cell from an initial population of cells. Furthermore, single-cell analysis has permitted to identify three subpopulations of ES cells, based on differences in chromatin signatures of pluripotency and differentiation priming, corresponding to cells belonging to the same cell line characterized by three different states of differentiation.
Rotem and collaborators [1] integrated alternative technologies with classical ChIP-seq, due to issues that interfere with the application of this method on single cells. In fact, ChIP-seq requires large amounts of input material and leads to experimental noise due to the binding of low levels of nonspecific antibodies to off-target sites; moreover, the quantity of on-target epitope is too low, especially in small-input experiments. These limitations were solved by using the microfluidics system, a technology based on the labelling, with different barcodes, of fragments obtained by MNase of the chromatin belonging to different cells. Drop-ChIP is a strategy for quickly profiling thousands of individual cells: the parallel analysis has been possible by encapsulating them in tiny droplets[2]. The device produces microdrops which contain a single cell . IN another droplet there is the buffer and enzymes and a thgird one contains a barcode library. A three points merging device allows to mix, once an electric field is applied, a unique barcode, with a single cell and an enzymatic buffer with DNA ligase. Each fragment belonging to a defined cell is marked on both its ends with a unique barcode: in this way is possible to combine chromatin from multiple cells before the immunoprecipitation step and recognize the originating cell of each fragment after the immunoprecipitation.
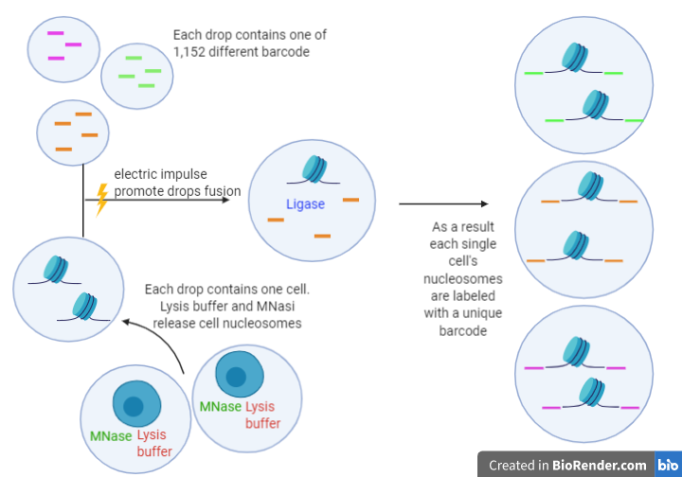
Schematic rapresentation of single cell barcoding with drop-based microfluidics system
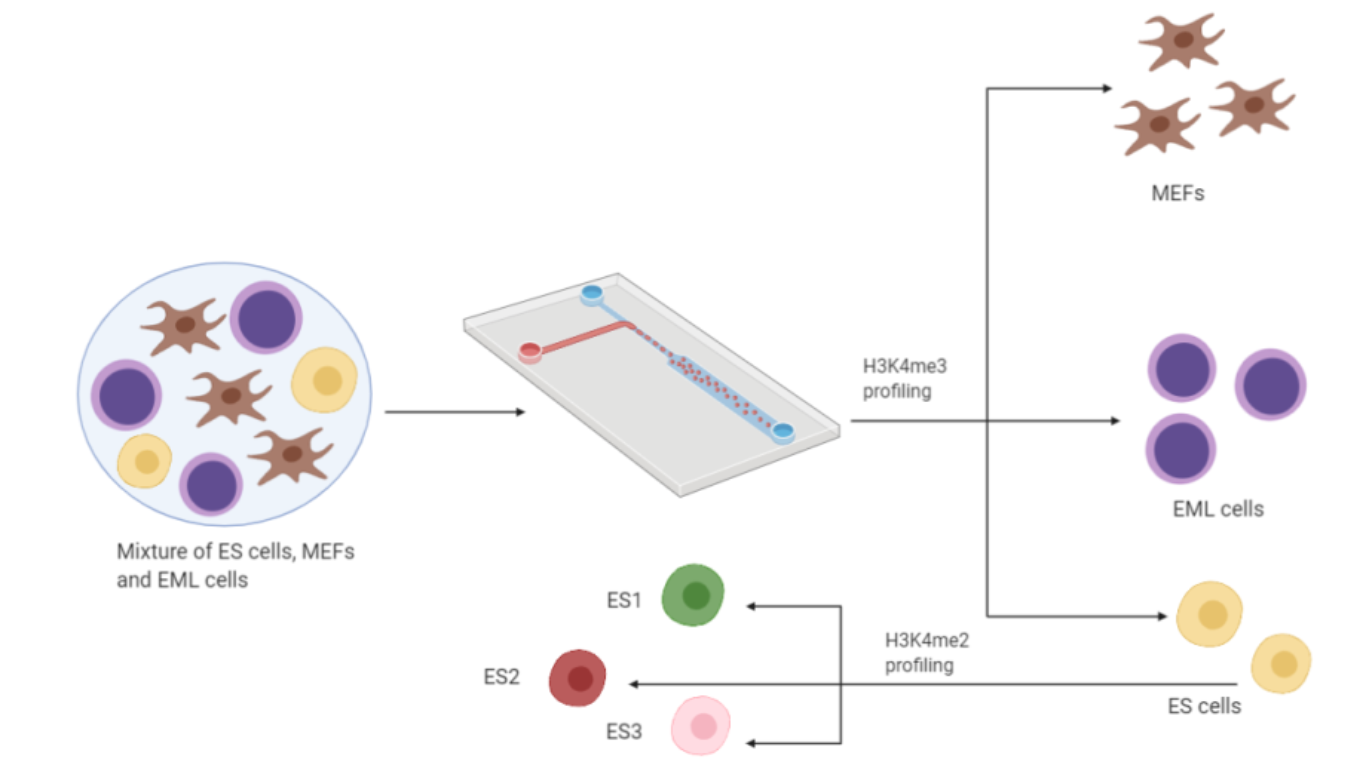
In the first part of the experiment Drop-ChIP with H3K4me3 antibody is performed in mixed populations of ES cells, MEFs and EML cells and the resulting library is sequenced. The comparison between single-cell data for 50 individual ES cells and 50 individual MEFs and data obtained from bulk ChIP-seq and aggregation of 200 profiles of single cells demonstrates the good quality of the experiment’s results. Additionally, the specificity of the differentially marked regions in different cell lines is enough to discriminate single-cell profiles for ES cells from single-cell MEF profiles. In fact, since epigenome differs from any cell type, it allows to instruct the unique gene expression program in each cytotype according to its genome [3]. A hierarchical agglomerative clustering (DIANA) identifies groups of related single cell profiles. Since three different sets of barcodes were used to label the three cellular lines, it’s possible to evaluate the accuracy of the clustering, which turns out to be very high (around 95%).
The other purpose for applying ChIP-seq at single cell resolution is discriminate differences among cells from the same population. In order to study this variability, the authors needed to increase signal-to-noise ratio: by analysing public database of genetic profiles and clustering functionally related genomic elements they indentified 91 signatures. Dimethylation of lysine 4 of histone 3 (H3K4me2) profile of ES cells and MEFs was analysed. As a results MEF cells appeared quite homogeneous, while among ES cells three different clusters could be identified. The three ESCs clusters were composed by a similar cells number and were called ES1, ES2 and ES3.
At this point, the authors investigated the biological significance of the ES subpolulations. Signatures which vary between ES1, ES2, and ES3 were analysed. In pluripotency-related signatures (as Oct4 and Sox2) ES1 had the higher signal, while ES2 signal was intermediate and ES3 signal was lower. An opposite signal distribution (higher signal in ES3, intermediate signal in ES2 and lower signal in ES1) was observed in FoxA2, CoREST, and Polycomb signatures. FoxA2 is an endodermal transcription factor which has a role in early ES cell differentiation. CoREST and Polycomb signatures are related to repressors and correlate inversely with pluripotency. In conclusion the ES cell subpopulations represent a different pluripotency degree, which is higher in ES1 and gradually lower in ES2 and ES3. ES3 also shows a less tight distribution, potentially reflecting an alternate priming state.
In conclusion, by using drop based microfluidics and DNA barcoding is possible to study chromatin state at single-cells resolution. Drop-ChIP-seq results to be able to identify cell type with high accuracy: the importance of this method is that it allows to study single cell variability among cell types. The experiment reported allowed to identify ES cells subpopulations which differ in pluripotency associated signals. By improving the single cell ChIP-seq method, it’s possible to better investigate correlation between chromatin state and gene expression which could be relevant to better understand regulatory elements and epigenetic mutation. In fact, these features can be mutated in diseases such as cancer [3] and a better understanding of these mechanism may lead to better treatment [4].
References
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. (2015) Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 33(11):1165-72
- Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., … & Trombetta, J. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell, 161(5), 1202-1214.
- Rivera, C. M., & Ren, B. (2013). Mapping human epigenomes. Cell, 155(1), 39-55.
- Baylin, S.B. & Jones, P.A. A decade of exploring the cancer epigenome–biological and translational implications. Rev. Cancer 11, 726–734 (2011)
- Grosselin K., Durand A., Marsolier J., Poitou A., Marangoni E., Nemati F., et al. (2019). High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Genet.51 1060–1066. 10.1038/s41588-019-0424-429