Is the analysis of liquid liquid phase separated chromatin useful to study in vitro the complex in vivo chromatin organization?
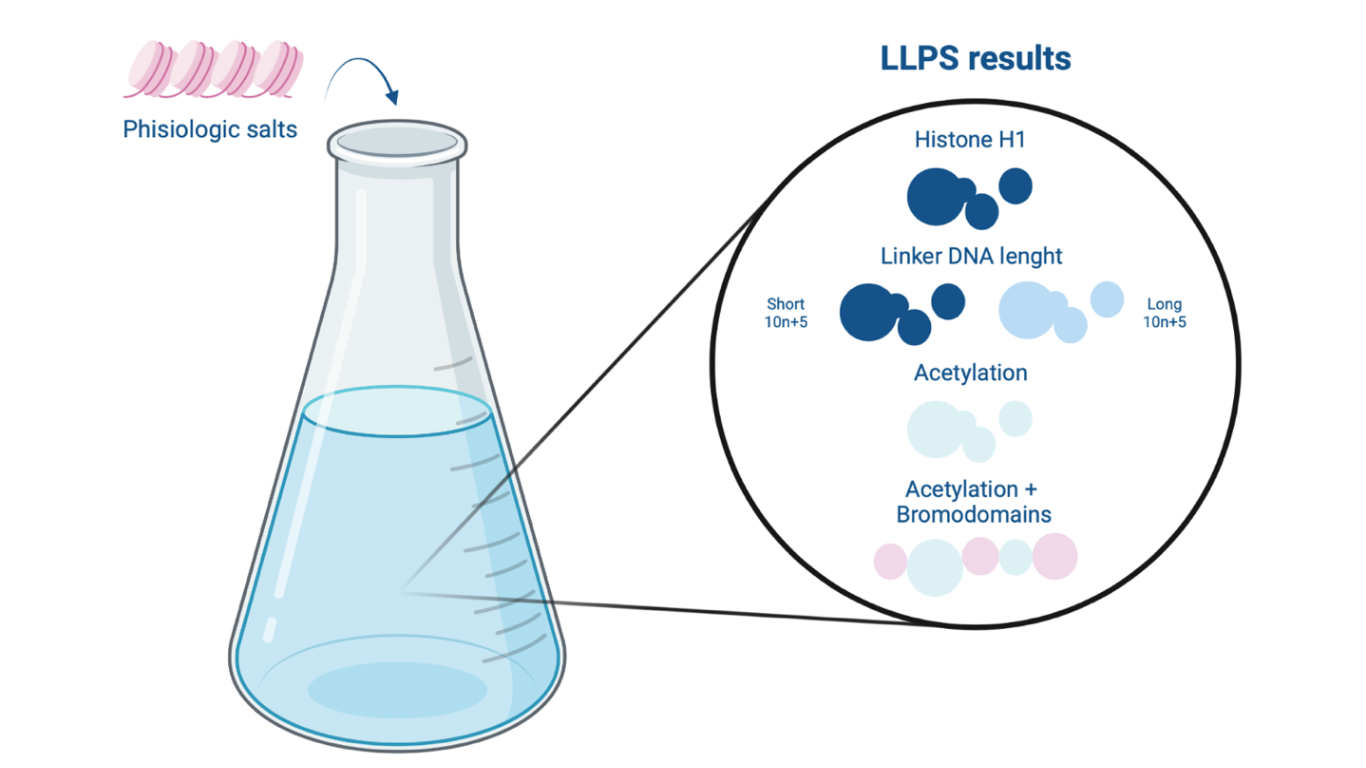
Figure 1 –Liquid-liquid phase separation (LLPS) as a results of different regulatory factors effect: linker histone H1, liker DNA length, acetylation and bromodomains.
Abstract
Can complex chromatin organization and its dynamicity be reproduced in vitro? Dr. Rosen Lab researchers have been engaged in studying in vitro the organizational changes in chromatin following the action of various regulatory factors, including histone H1, internucleosomal spacing, acetylation and linker histone binding proteins [1]. By observing the LLPS phenomenon (Liquid-Liquid Phase Separation) they were able to study how the properties of the resulting droplets reflected the structural behaviors of chromatin in vivo, thus proposing a true phase separation-based model for studying chromatin organization.
Discussion
DNA is a large structure that must be arranged within a very small space, the nucleus. For this reason it is necessary for some portions of it to be more compact than others. As a consequence, differentiated cells make only the momentarily relaxed regions available to the transcriptional machinery. The basic unit of DNA is the nucleosome: 146 bp wrapped around a histone octamer. The histones N-terminal tails are centers for major histone modifications (acetylation, methylation, phosphorylation), the combination of which is aimed at making DNA more or less accessible 2.
With the aim of demonstrating the behavior that chromatin takes on in vivo, the authors proposed that the LLPS phenomenon can be involved in vitro to study morphological and organizational changes of chromatin following the action of different regulatory factors.
In vitro, various cations promote self-association of chromatin, resulting in its precipitation from solution 3. To reproduce the physiological conditions characterizing the internuclear space, the authors used a solution of mono- and divalent salts (KOAc, Mg[OAc]2). Chromatin sample containing 12 repeats of the Widom 601 nucleosome positioning sequence 4 and fluorophore-labeled histone octamers were then introduced into it. Taking advantage of fluorescence confocal microscopy, it was possible to detect the formation of phase-separated chromatin droplets. This phenomenon can be attributed to the LLPS phenomenon. The dynamics of the droplets obtained as a result of LLPS can be assessed by photobleaching, a technique used to study the mobility and diffusion of fluorophore-labelled molecules following the photochemical alteration of the fluorophore 5: on the basis of the time it takes for the droplet to recover fluorescence, its dynamics can be determined. The study of fluorescence is also useful for measuring the density of droplets: as the fluorescence intensity of chromatin droplets increases, their density increases. The increase in dynamism and decrease in chromatin droplet density signify a decrease in DNA compaction, thus corresponding to more accessible chromatin.
What effect do regulatory factors have on the morphology and dynamism of chromatin droplets in vitro?
The first regulatory factor considered is the histone linker H1: it represents the most abundant structural chromatin-binding protein in eukaryotes. Its main task is to regulate genomic condensation, consequently transcriptional machinery access, by interacting with both internucleosome spacing and the nucleosome 6. The presence of histone H1 in the chromatin structure promotes in vitro the formation of very dense and low-dynamic droplets reflecting the function of the histone linker in vivo to condense chromatin.
The study then analyzed the role of internucleosome spacing, the portion of DNA interposed between two nucleosomes. All eukaryotes are characterized by a 10n+5 internucleosome spacing, which length increases as the complexity of the organism increases. Shorter nucleosome spacing generate more dense and less dynamic chromatin droplets, on the other hand longer spacing give rise to less dense and more dynamic droplets. Correlating the internucleosome length with the presence of the linker histone H1, it was seen that simpler eukaryotes require less histone H1 because they achieve sufficient levels of chromatin compaction with shorter internucleosome spacing however more complex eukaryotes require more histone H1 because they possess longer internucleosome spacing 7.
Next, researchers examined how acetylation of histone tails, conducted by histone-acetyltransferase enzymes in vitro, can alter the properties of chromatin droplets with a decreasement in their density. It was also demonstrated how, at high levels of chromatin acetylation, droplets do not appear. This shows how, in vivo, acetylation of histone tails can give rise to relaxed chromatin.
It is well known how highly acetylated histone tails, thus transcriptionally active genomic sites, can be recognized by bromodomains, a bundle of four helices characterizing numerous chromatin-associated proteins with remodeling function 8. The study wanted to demonstrate how the addition of multi-bromodomain proteins (BRD4 and bromo5) into an acetylated chromatin solution enabled phase separation, thus the formation of droplets with different properties (less dynamic and less dense than droplets of unacetylated chromatin). Bromo5-guided LLPS demonstrates how, in vivo, acetylated chromatin regions are spatially separated from nonacetylated chromatin regions. The chromatin droplets, in fact, interact without fusing.
Conclusions
The authors, through this study, aimed to show that the LLPS phenomenon observable in vitro is actually what occurs in vivo as well: chromatin within the nucleus undergoes phase separation driven by regulatory factors by dividing into euchromatin and heterochromatin.
In cells, there are different defined functional chromatin subtypes (e.g., promoters, enhancers, insulators, Polycomb group regions, etc…). Just as it has been possible to exploit phase separation to study the organization and arrangement of chromatin within the nucleus, based on histone modifications and regulatory factors acting on euchromatin, this study aims to demonstrate how each euchromatin subtype can adopt a different phase separation depending on the type of modification it undergoes. For example, further studies analyzed how some particular proteins, containing IDRs (intrinsically disordered regions), give rise to compartments of dense chromatin organized through liquid-liquid phase separation [See the Article – Disordered regions in transcription factors lead to short-order binding].
The researchers thus assume that the chromatin phase separation analyzed so far is in a “basal” state, at the same time they wonder whether it is possible to generate different “excited” structural states demonstrating how each euchromatin subtypes can adopt a different phase separation depending on the type of histone modification it undergoes (acetylation, methylation, phosphorylation, ubiquitination) and the types of regulatory proteins it binds on.
The question is: is LLPS phenomenon really useful to investigate the generation of “excited” structural states? Isn’t too artificial the fact that they want to make droplets of euchromatin subtypes appear in vitro? Phase separation of euchromatin subtypes is not only driven by chromatin, but also by the proteins bound to it, the histone code and the nucleotide sequence. Perhaps it could be very complex to differentiate and to establish the properties of the many euchromatin droplets that would be obtained.
References
- Gibson et al., 2019, Cell 179, 470–484 October 3, 2019 a 2019 Elsevier Inc. https://doi.org/10.1016/j.cell.2019.08.037
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001; 293: 1074-1080
- Hansen J.C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002; 31: 361-392
- Spakman, D., King, G.A., Peterman, E.J.G. et al. Constructing arrays of nucleosome positioning sequences using Gibson Assembly for single-molecule studies. Sci Rep 10, 9903 (2020).
- Williamson DE, Sahai E, Jenkins RP, O’Dea RD, King JR. Parameter estimation in fluorescence recovery after photobleaching: quantitative analysis of protein binding reactions and diffusion. J Math Biol. 2021;83(1):1. Published 2021 Jun 15. doi:10.1007/s00285-021-01616-z
- Bednar, et al. (2017). Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Mol. Cell 66, 384–397.e8.
- Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006; 14: 17-25
- Fujisawa T., Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer.