Controlling intestinal organoid regeneration with retinoic acid and RXR inhibition
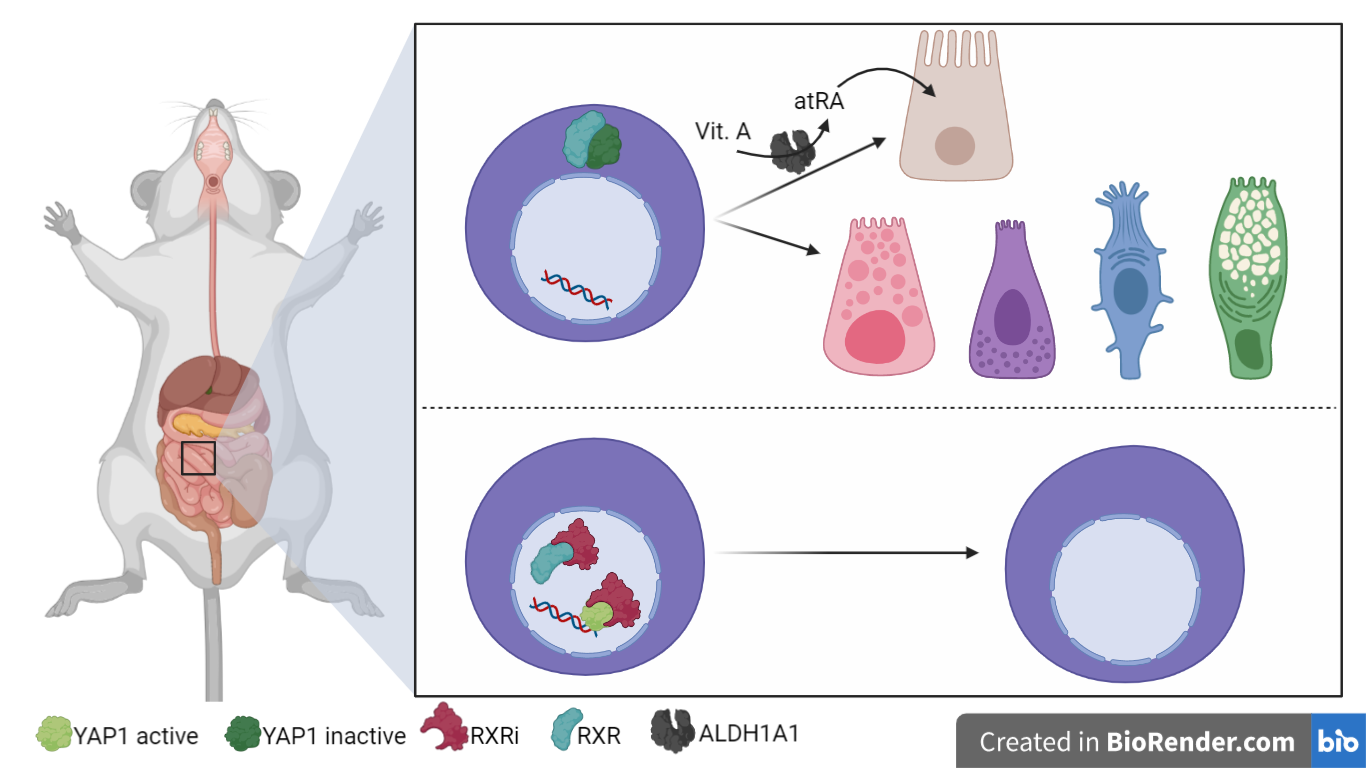
Fig. 1 – RXR is a homing mechanism to exit regenerative state and its inhibition leads to regenerative phenotype; retinoic acid provides tissue-specificity [Created with BioRender.com]
Abstract
Discussion
An entire organoid develops from a single cell, forming a self-organized structure. Initially, single cells enter a regenerative state that is dependent on the transcriptional regulator YAP, and forms a symmetric cyst [2]. Subsequently, symmetry is broken by the emergence of Paneth cells that define and maintain the crypt, followed by the differentiation of absorptive enterocytes distal from the crypt [3]. Organoids thus recapitulate the regeneration of the epithelium and subsequent re-establishment of homeostasis [4]. In this work [5], the authors established an image-based screening platform to characterize the phenotypic landscape of organoid development from a single screen, generating the first map of functional genetic interactions that govern intestinal organoid development and self-organization.
Organoids development and screening
RXR inhibition leads to regenerative phenotype
Next, the authors focused on the regenerative phenotype. During this phase, they noticed that the use of the RXR antagonist Cpd2170 (RXRi) [7] led to an increase in the regenerative phenotype. In particular, the authors measured the abundance of enterocytes through aldolaseB staining and noticed that the antagonist of RXR induces a near-complete absence of enterocytes. In contrast, they observed that an RXR agonist, all-trans retinoic acid (atRA), led to the opposite effect. In fact, it increased enterocyte differentiation despite failing to rescue the RXRi-induced differentiation defect, hinting at a retinoic acid-independent role of RXR.
To investigate the lack of Paneth cells that was also observed upon RXR inhibition, the authors turned to Notch signaling and YAP1 localization. In RXRi-treated organoids they observed an absence of cells expressing the Notch ligand DLL1 and a strong homogenous nuclear retention of YAP1, which led to retaining a regenerative state [2] and an active cell cycle. By contrast, in atRA-treated organoids YAP1 was invariably localized to the cytoplasm and cells underwent differentiation to enterocytes. However, both RXRi-and atRA-treated organoids showed a defect in symmetry breaking. Interestingly, RXRi-treatment after YAP1 activation did not result in nuclear translocation of YAP1, suggesting that RXR does not activate YAP1 but rather controls the nuclear export.
All-trans retinoic acid role
ALDH1A1 knock-out of this gene or vitamin A depletion led to fewer enterocytes and more cycling cells, suggesting that retinoic acid metabolism is necessary for differentiation of enterocytes but not of Paneth cells.
In vivo mice experiment
By treating mice with RXRi they observed improved barrier function and increasingly significant villi length recovery as days passed but never reached initial normal size.
In addition, the treatment improved crypt morphology by accumulating proliferative cells at the bottom. These results confirmed the regenerative phenotype also in vivo, suggesting RXRi treatment as a possible useful therapy to improve intestine regeneration.
Conclusions
The interaction between RXRi and retinoic acid could be used to obtain new experimental models in vitro by manipulating the intestinal gene pool. In the future, this regeneration potential could be explored by combining inhibitors with different diets. In addition, inhibition could be used to mitigate problems that may arise with cancer therapies that damage the intestinal barrier by affecting stem cells
References
- Garabedian, E. M., Roberts, L. J. J., McNevin, M. S., & Gordon, J. I. (1997). Examining the Role of Paneth Cells in the Small Intestine by Lineage Ablation in Transgenic Mice. Journal of Biological Chemistry, 272(38), 23729–23740.
- Serra, D., Mayr, U., Boni, A., Lukonin, I., Rempfler, M., Challet Meylan, L., Stadler, M. B., Strnad, P., Papasaikas, P., Vischi, D., Waldt, A., Roma, G., & Liberali, P. (2019). Self-organization and symmetry breaking in intestinal organoid development. Nature, 569(7754), 66–72.
- Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., van Es, J. H., Abo, A., Kujala, P., Peters, P. J., & Clevers, H. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature, 459(7244), 262–265.
- Chacón-Martínez, C. A., Koester, J., & Wickström, S. A. (2018). Signaling in the stem cell niche: regulating cell fate, function and plasticity. Development, 145(15).
- Lukonin, I., Serra, D., Challet Meylan, L., Volkmann, K., Baaten, J., Zhao, R., Meeusen, S., Colman, K., Maurer, F., Stadler, M. B., Jenkins, J., & Liberali, P. (2020). Phenotypic landscape of intestinal organoid regeneration. Nature, 586(7828), 275–280.
- Snijder, B., Liberali, P., Frechin, M., Stoeger, T., & Pelkmans, L. (2013). Predicting functional gene interactions with the hierarchical interaction score. Nature Methods, 10(11), 1089–1092.
- Sakaki, J., Kishida, M., Konishi, K., Gunji, H., Toyao, A., Matsumoto, Y., Kanazawa, T., Uchiyama, H., Fukaya, H., Mitani, H., Arai, Y., & Kimura, M. (2007). Synthesis and structure–activity relationship of novel RXR antagonists: Orally active anti-diabetic and anti-obesity agents. Bioorganic & Medicinal Chemistry Letters, 17(17), 4804–4807.
- Haber, A. L., Biton, M., Rogel, N., Herbst, R. H., Shekhar, K., Smillie, C., Burgin, G., Delorey, T. M., Howitt, M. R., Katz, Y., Tirosh, I., Beyaz, S., Dionne, D., Zhang, M., Raychowdhury, R., Garrett, W. S., Rozenblatt-Rosen, O., Shi, H. N., Yilmaz, O., … Regev, A. (2017). A single-cell survey of the small intestinal epithelium. Nature, 551(7680), 333–339.