Genetic engineering of human organoids using a homology independent CRISPR-Cas9
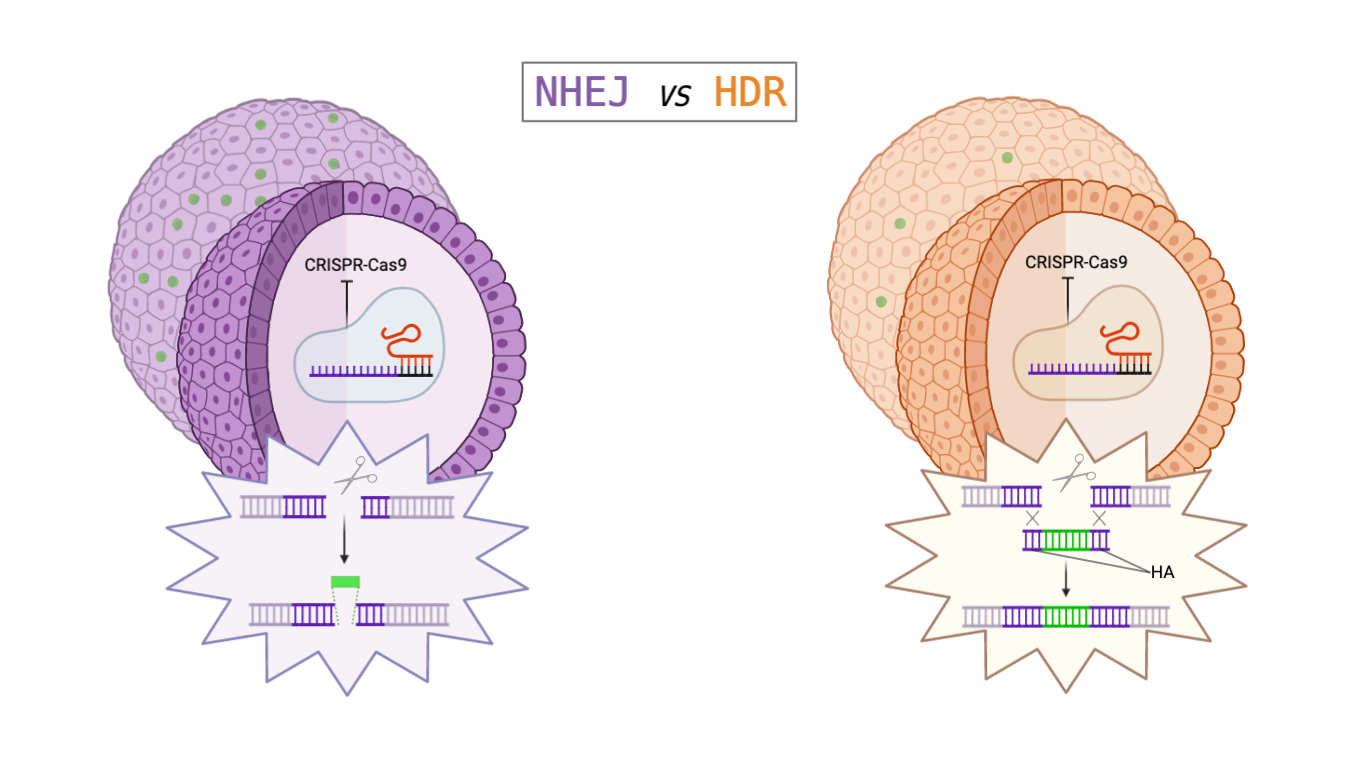
Figure 1 – Comparison between HDR and NHEJ mechanism. In the HDR mechanism are highlighted the homology arms (HA). Created with Biorender.
Abstract
Organoids are in vitro generated miniaturized versions of organs that simulate the structure and function of the corresponding organ in vivo. Engineering these systems’ genomes can be performed using CRISPR-Cas9, but the efficiency of the technique is affected by the DNA repair mechanism used by the cell. Most known DNA repair mechanisms are Homology Directed Repair (HDR) and Non-Homologous End-Joining (NHEJ); here Benedetta Artegiani et al. show that NHEJ provides a faster and more efficient gene editing, compared to other methods. The novel technique, called CRISPR-Cas9-mediated Homology-independent Organoid Transgenesis (CRISPR–HOT) was used to successfully generate human intestinal and hepatocyte organoid reporter lines. These latter lines were used to analyse mitotic spindle dynamics and cell division. The results demonstrated different unconventional hepatocytes divisions, characterised by a polypoid state maintenance and a correlation between mitotic spindle orientation and cell polarity. Summing up, CRISPR-HOT simplifies gene editing in human organoids and represents a valuable resource for future studies.
Discussion
In recent years, biotechnology research has focused on developing in vitro biological systems that faithfully reproduce the same conditions of cells in vivo.
This aim has been achieved by generating a miniaturised form of organs, called organoids. Organoids can be applied in many fields, like toxicology, drug discovery, disease modelling and gene editing. Many strategies were developed to edit the genome, in order to provide both knock-in (KI) and knock-out (KO); CRISPR-Cas9 is the newest one, discovered a few years ago as an adaptive immune system of some bacteria against viral infection. From this discovery, CRISPR-Cas9 found broad applications as programmable nucleases for the disruption, the deletion, or the insertion of genes and it has been discovered to solve most of the problems of the previously used methods. However, knock-in still remains a big challenge for many applications. In this article, Artegiani et al [1] presented a novel, fast and efficient method.
CRISPR-Cas9 and DNA repair mechanisms
CRISPR-Cas9 can be seen as a molecular scissor composed of a Cas9 and a small guide RNA sequence. Cas9 is an endonuclease that is able to induce a double strand break (DSB) in a specific position of the DNA, thanks to its ability to bind the small guide RNA, which is complementary to the target DNA region. The small guide RNA itself consists of crispr-RNA (crRNA), a 17-20 nucleotide sequence complementary to the target DNA, and of tracr-RNA (trRNA), which serves as a binding scaffold for the Cas nuclease [2]. After DSBs, two different mechanisms can occur in the cell to repair the double strand: the Homology Directed Repair (HDR) and the Non-Homologous End-Joining (NHEJ) (Figure 1) [3].
The HDR needs two homology arms flanking the region of interest of the gene where insertion has to occur. This mechanism occurs only in a specific cell cycle phase (the S phase) and is not very efficient, so it is not the best option to achieve knock-in. HDR efficiency has been improved in previous strategies by deleting TP53 gene, in order to reduce the DNA damage response and to avoid the consequent transient cell-cycle arrest [4].
The NHEJ mechanism, instead, is independent from the cell cycle phases and does not require the time-consuming homology arms cloning. By the way it is widely considered as an error prone mechanism since it involves the addition or deletion of random nucleotides to join the two ends.
CRISPR-HOT: Development and validation
In this work, Benedetta Artegiani et al. developed a method that combines the DSB induced by CRISPR-Cas9 and the NHEJ repair mechanism, in order to obtain a fast and efficient knock-in in human liver and intestinal organoids.
Efficiency evaluation
Firstly, was compared the knock-in efficiency between the HDR and NHEJ as repair mechanism. The KI was performed by targeting two different loci (KRT19 for ductal hepatocytes and TUBB for hepatocytes) with mNEON and tdTomato genes which encode for two different fluorescent proteins. Knock-in efficiency was evaluated using Fluorescence-Activated Cell Sorting (FACS) technology, that uses a laser to detect the fluorescent protein when transgenesis is properly achieved.
The results showed that the NHEJ method provides a higher knock-in efficiency than HDR, even when this is performed in cells expressing a dominant negative form of TP53 (DNTP53), mimicking the TP53 KO phenotype.
Precision evaluation
In addition, the knock-in precision was evaluated. For this aim, TUBB and CDH1 genes were targeted in human liver organoids. Knocked-in cells were assessed using FACS and the genotype was analysed by sequencing.
The results showed that 30% of the cells had an in-frame insertion, while the remaining cells showed no insertions or out-of-frame insertions.
Overall, both rates of precision and efficiency confirmed the robustness of the technology which was finally re-nominated as CRISPR-HOT (CRISPR-Cas9-mediated Homology-independent Organoid Transgenesis).
Generation of human reporter organoids lines
Once CRISPR-HOT was confirmed to be a precise and efficient method to knock in cells, it was used to label rare intestinal cells (enteroendocrine and goblet cells), and to generate human reporter organoid lines. In particular, liver ductal and hepatocyte reporter organoid lines were generated by targeting CDH1 and TUBB genes. All the derived lines presented precise in-frame knock-in, confirming the validity of CRISPR–HOT.
Mitotic spindle and hepatocytes division analysis
The human liver ductal reporter lines were then used to study the mitotic spindle dynamics and hepatocyte division. A double knocked-in TUBB::mNEON; CDH1::tdTomato clonal line was generated to visualise simultaneously the cell membrane (with the tdTomato tagging the E-cadherin protein) and the mitotic spindle (with the mNeon tagging the tubulin).
From this analysis emerged that hepatocytes dividing next to a lumen undergo symmetric division with respect to the apicobasal axis. Interestingly, the authors found that binucleated cells (that are very common in liver) maintained their polyploid state after unconventional division.
Consequences of TP53 knock-out
Afterwards CRISPR–HOT was used to perform TP53 knock-out to study the effect of TP53 loss in human hepatocyte reporter lines, because it is well known that TP53 is involved in genomic stability. TP53 Knocked-out hepatocytes showed the formation of non-canonical mitotic spindles and an increase in the polyploid state (3-4 nuclei), though retaining the ability to divide.
Conclusions
In this work it is shown that CRISPR-HOT is a homology-independent strategy that can be used to achieve fast, efficient, and precise gene knock-in, outperforming the HDR method, even when this is combined with transient TP53 loss.
CRISPR-HOT allowed to create with high efficiency and precision organoid reporter lines by targeting both constitutively and non-constitutively expressed genes as shown for hepatocytes and rare intestinal cells.
This study also demonstrated that there is a correlation between mitotic spindle orientation and tissue polarity; in addition, the typical polyploid state of hepatocytes is maintained, after division process, and increased with the loss of TP53 gene.
Future perspectives
To sum up, in the future CRISPR-HOT could be applied as a useful tool in studies that require the generation of reporter lines, protein tagging, labelling of cellular structures and lineage tracing experiments.
Then, CRISPR-HOT could find application to other human cell models, such as organoids derived from induced pluripotent stem cells and embryonic stem cells.
References
- Benedetta Artegiani et al, «Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing,» Nature Cell Biology, vol. 22, pp. 321-331, 2020.
- Ekaterina Kondrateva et al, «An overview of currently available molecular Cas-tools for precise genome modification,» Gene, vol. 769, 2021.
- Juliët Schreurs et al, «Recent Advances in CRISPR/Cas9-Based Genome Editing Tools for Cardiac Diseases,» International Journal of Molecular Sciences, vol. 22, 2021.
- Emma Haapaniemi et al, « CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response,» Nature Medicine, vol. 24, p. 927–930, 2018.